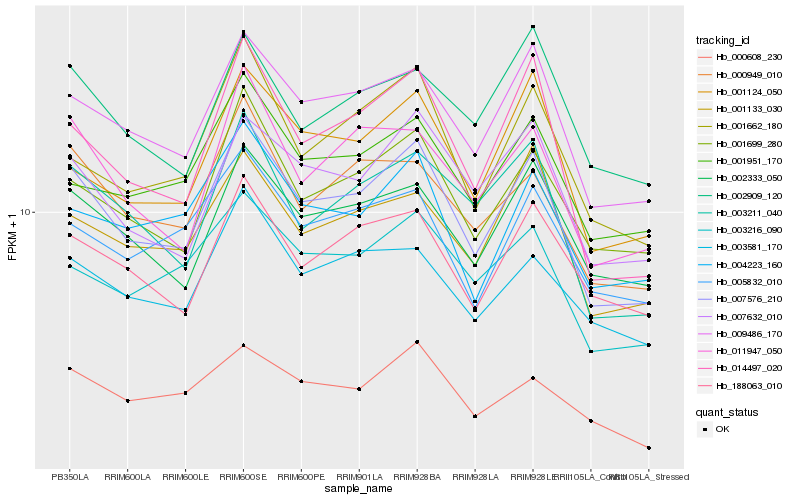
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_210 |
0.0 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 2 |
Hb_009486_170 |
0.0565079689 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 3 |
Hb_014497_020 |
0.0654435147 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 4 |
Hb_001699_280 |
0.0694005111 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 5 |
Hb_007632_010 |
0.0708788087 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002333_050 |
0.0714726212 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003211_040 |
0.0717687631 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000949_010 |
0.0739271952 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 9 |
Hb_011947_050 |
0.0739618726 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 10 |
Hb_001133_030 |
0.0775998587 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 11 |
Hb_188063_010 |
0.0781076208 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 12 |
Hb_001124_050 |
0.0785055768 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 13 |
Hb_003216_090 |
0.0787629798 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001662_180 |
0.0791027606 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 15 |
Hb_004223_160 |
0.0820633618 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003581_170 |
0.083831251 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005832_010 |
0.0840801722 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001951_170 |
0.0853024431 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 19 |
Hb_000608_230 |
0.0858039239 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002909_120 |
0.086324254 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |