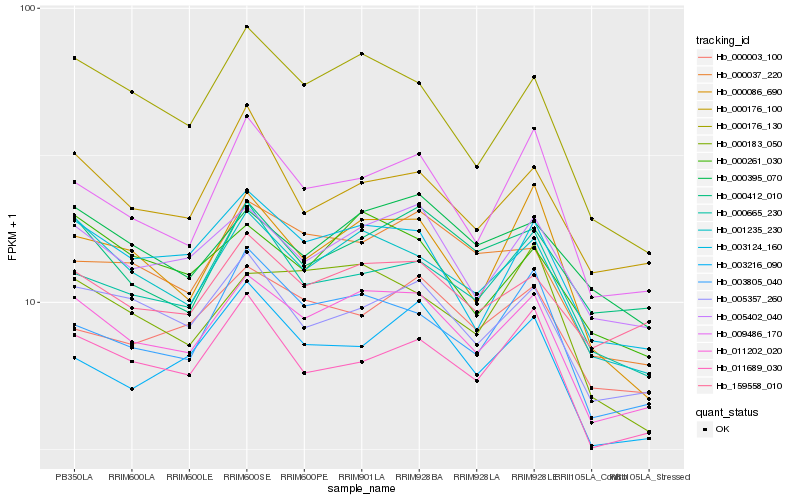
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011202_020 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001235_230 |
0.0461788901 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 3 |
Hb_005402_040 |
0.0534439587 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 4 |
Hb_003124_160 |
0.0587749544 |
- |
- |
dynamin, putative [Ricinus communis] |
| 5 |
Hb_005357_260 |
0.0600246091 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 6 |
Hb_000261_030 |
0.0654243929 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 7 |
Hb_009486_170 |
0.0658224377 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 8 |
Hb_000176_130 |
0.0665486404 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 9 |
Hb_159558_010 |
0.0667252996 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 10 |
Hb_003805_040 |
0.0669463118 |
- |
- |
Sacsin [Glycine soja] |
| 11 |
Hb_011689_030 |
0.0674318807 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 12 |
Hb_000665_230 |
0.0698815588 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_000037_220 |
0.0706745212 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 14 |
Hb_000176_100 |
0.071171364 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 15 |
Hb_000395_070 |
0.0712204681 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000183_050 |
0.073260015 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 17 |
Hb_000086_690 |
0.0737011522 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 18 |
Hb_000003_100 |
0.0738833394 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 19 |
Hb_000412_010 |
0.0746476369 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_003216_090 |
0.0757353891 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |