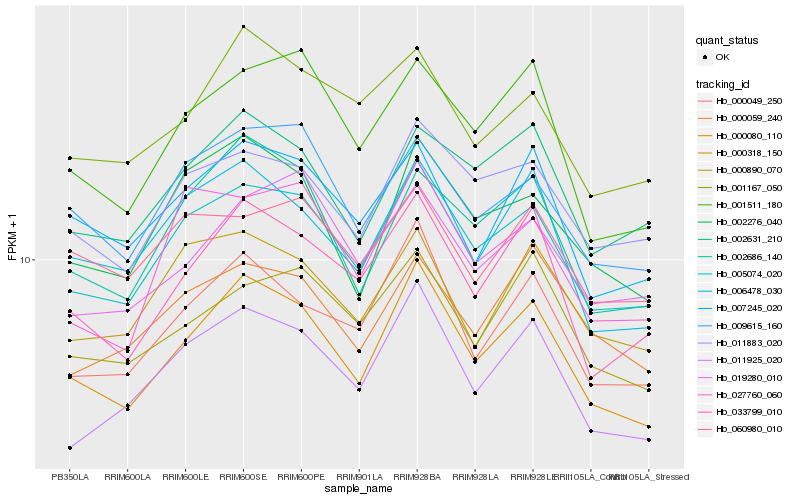
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005074_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011883_020 |
0.0534425208 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 3 |
Hb_000080_110 |
0.0571614462 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_027760_060 |
0.0680205386 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 5 |
Hb_009615_160 |
0.0686902212 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 6 |
Hb_002276_040 |
0.0699167945 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 7 |
Hb_011925_020 |
0.0705651367 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_002631_210 |
0.0720451687 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 9 |
Hb_000049_250 |
0.0729730359 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001511_180 |
0.0732222839 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 11 |
Hb_000318_150 |
0.0748733671 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 12 |
Hb_033799_010 |
0.0759674421 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 13 |
Hb_019280_010 |
0.0774453319 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006478_030 |
0.07751206 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 15 |
Hb_002686_140 |
0.078839432 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_060980_010 |
0.0792191009 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000890_070 |
0.0793439894 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001167_050 |
0.0807388068 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 19 |
Hb_000059_240 |
0.0807907505 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 20 |
Hb_007245_020 |
0.0811469288 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |