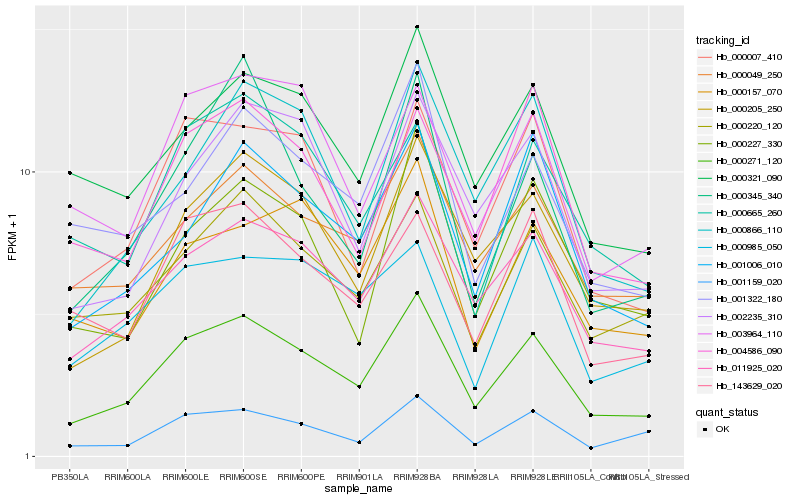
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000271_120 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 2 |
Hb_011925_020 |
0.0733766731 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_000205_250 |
0.0838580956 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 4 |
Hb_000007_410 |
0.0911506157 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 5 |
Hb_000049_250 |
0.0922590695 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000220_120 |
0.0960393222 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001006_010 |
0.0965438452 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 8 |
Hb_002235_310 |
0.098774518 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 9 |
Hb_000866_110 |
0.1012705971 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 10 |
Hb_143629_020 |
0.1022563814 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 11 |
Hb_000321_090 |
0.1024550799 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 12 |
Hb_001159_020 |
0.1033803982 |
- |
- |
- |
| 13 |
Hb_000227_330 |
0.1063194981 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 14 |
Hb_000157_070 |
0.1074804746 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 15 |
Hb_001322_180 |
0.1093207348 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 16 |
Hb_004586_090 |
0.1096300598 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000345_340 |
0.1114573251 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 18 |
Hb_003964_110 |
0.1120160208 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000665_260 |
0.115156022 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000985_050 |
0.1158058587 |
- |
- |
PREDICTED: uncharacterized protein LOC105647985 isoform X1 [Jatropha curcas] |