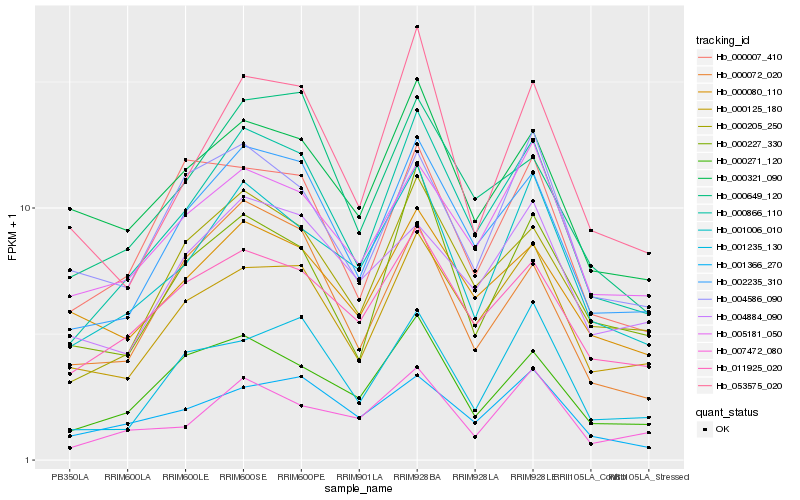
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002235_310 |
0.05294252 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 3 |
Hb_000205_250 |
0.0743171539 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 4 |
Hb_000125_180 |
0.0866862607 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 5 |
Hb_001006_010 |
0.0882424942 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 6 |
Hb_011925_020 |
0.0941365471 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_000271_120 |
0.1012705971 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 8 |
Hb_000649_120 |
0.1012783008 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_005181_050 |
0.1074638235 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 10 |
Hb_053575_020 |
0.1075705858 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 11 |
Hb_001366_270 |
0.1079428693 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 12 |
Hb_000227_330 |
0.1094715724 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 13 |
Hb_001235_130 |
0.1166567896 |
- |
- |
- |
| 14 |
Hb_007472_080 |
0.1172241941 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 15 |
Hb_000080_110 |
0.117484168 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000321_090 |
0.1180617774 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 17 |
Hb_000007_410 |
0.1192537148 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 18 |
Hb_000072_020 |
0.119861039 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004884_090 |
0.1232697318 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 20 |
Hb_004586_090 |
0.1243058841 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |