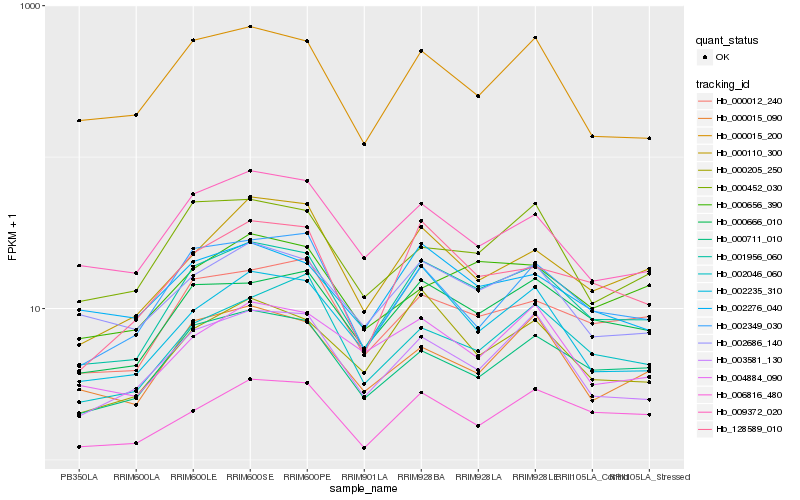
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001956_060 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 2 |
Hb_000666_010 |
0.0776488635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000012_240 |
0.0896103614 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000711_010 |
0.0903969883 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 5 |
Hb_128589_010 |
0.0941603003 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 6 |
Hb_003581_130 |
0.1008453769 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000110_300 |
0.1009448621 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 8 |
Hb_002686_140 |
0.1010323092 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002349_030 |
0.1023891217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000205_250 |
0.1040792453 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 11 |
Hb_000015_200 |
0.1044042796 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 12 |
Hb_009372_020 |
0.1056022633 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 13 |
Hb_002235_310 |
0.1082014232 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 14 |
Hb_000015_090 |
0.1126198076 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 15 |
Hb_002046_060 |
0.1126672835 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 16 |
Hb_002276_040 |
0.1137141772 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 17 |
Hb_006816_480 |
0.1150195143 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004884_090 |
0.1154555399 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 19 |
Hb_000452_030 |
0.1157372465 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_000656_390 |
0.1159443729 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |