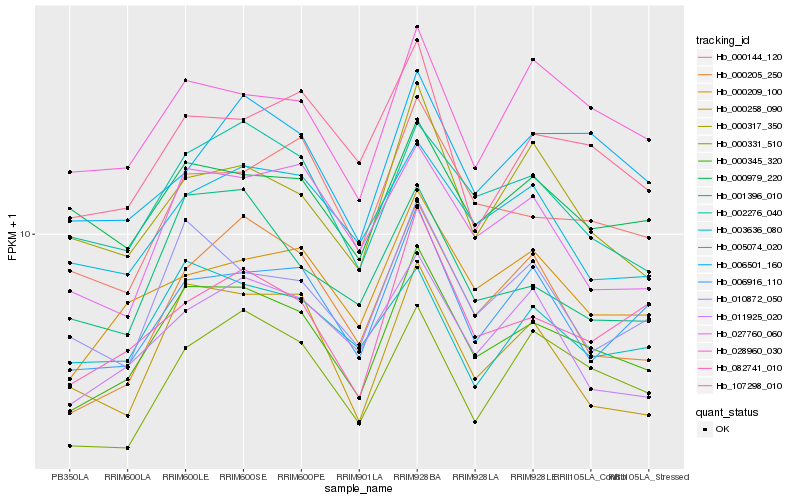
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_320 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002276_040 |
0.098049778 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 3 |
Hb_082741_010 |
0.1010458928 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 4 |
Hb_000209_100 |
0.1018799616 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006916_110 |
0.1022177377 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010872_050 |
0.1023623199 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 7 |
Hb_000144_120 |
0.1040694606 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_028960_030 |
0.1050426903 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 9 |
Hb_001396_010 |
0.1072265953 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_027760_060 |
0.1093821961 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_107298_010 |
0.1120258541 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 12 |
Hb_000205_250 |
0.1136562517 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 13 |
Hb_000317_350 |
0.1141048996 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 14 |
Hb_011925_020 |
0.1143723589 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_003636_080 |
0.1143989656 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 16 |
Hb_005074_020 |
0.1159738641 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000331_510 |
0.1163210992 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 18 |
Hb_006501_160 |
0.116489956 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 19 |
Hb_000979_220 |
0.1165517533 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000258_090 |
0.1169459075 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |