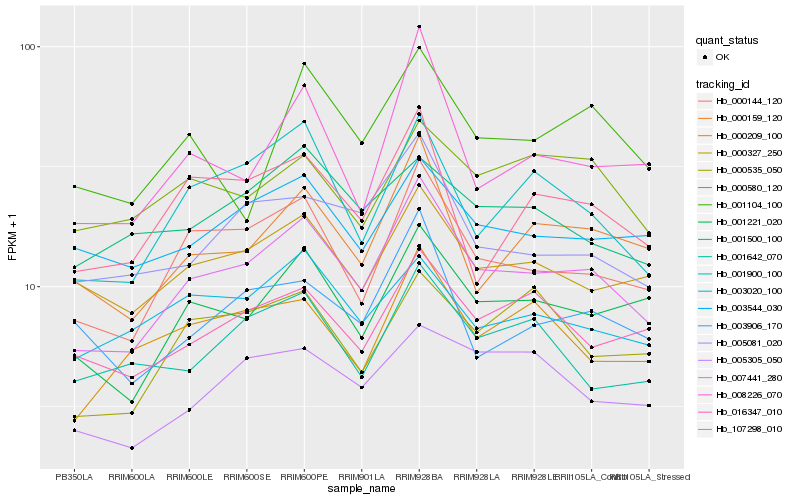
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_280 |
0.0 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 2 |
Hb_000144_120 |
0.0712744876 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_001221_020 |
0.0920402066 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 4 |
Hb_001900_100 |
0.0953129173 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000159_120 |
0.0971597039 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 6 |
Hb_005081_020 |
0.0988042088 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 7 |
Hb_000327_250 |
0.0999139106 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 8 |
Hb_107298_010 |
0.1052078635 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 9 |
Hb_016347_010 |
0.1073713523 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_008226_070 |
0.10741454 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 11 |
Hb_000209_100 |
0.1089951752 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003544_030 |
0.109119947 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 13 |
Hb_003906_170 |
0.1125347748 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_005305_050 |
0.1130685993 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 15 |
Hb_003020_100 |
0.1141095946 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 16 |
Hb_001642_070 |
0.1143691698 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 17 |
Hb_000535_050 |
0.1157166001 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 18 |
Hb_001500_100 |
0.1168392288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001104_100 |
0.117107032 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 20 |
Hb_000580_120 |
0.1182225221 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |