| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
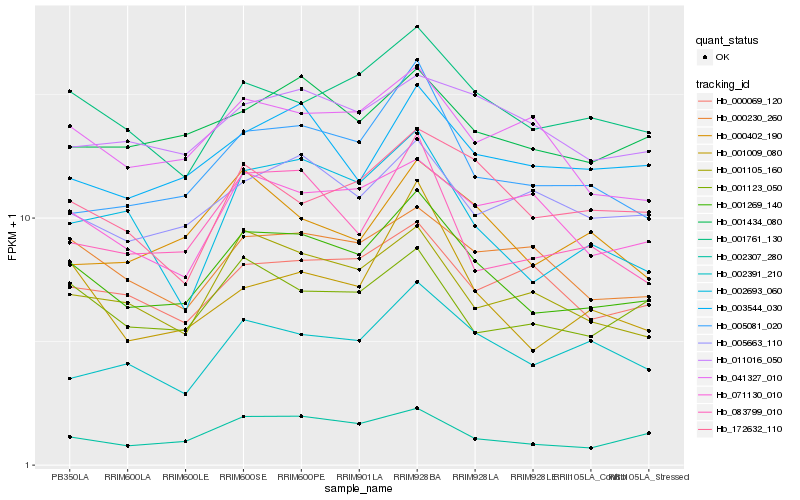
Hb_001269_140 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 2 |
Hb_002307_280 |
0.0774101895 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 3 |
Hb_001761_130 |
0.0851847935 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005081_020 |
0.0904616848 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 5 |
Hb_001105_160 |
0.0907816812 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 6 |
Hb_001434_080 |
0.0947027926 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 7 |
Hb_001123_050 |
0.0955566188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_083799_010 |
0.0977456117 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_041327_010 |
0.0980021676 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 10 |
Hb_000230_260 |
0.0985274015 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 11 |
Hb_002693_060 |
0.0989217304 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_003544_030 |
0.0995516958 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 13 |
Hb_005663_110 |
0.1029771907 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 14 |
Hb_071130_010 |
0.1035236189 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000069_120 |
0.1042454713 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 16 |
Hb_002391_210 |
0.1042713062 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 17 |
Hb_011016_050 |
0.1044210281 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_172632_110 |
0.1050804382 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 19 |
Hb_001009_080 |
0.1055250664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000402_190 |
0.1058680408 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |