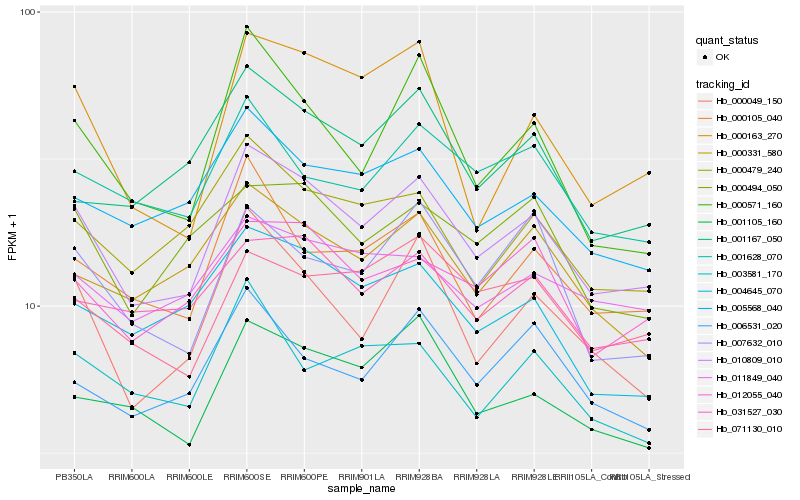
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010809_010 |
0.0 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 2 |
Hb_000105_040 |
0.0776258616 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 3 |
Hb_000049_150 |
0.0782003432 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_001105_160 |
0.080583292 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 5 |
Hb_005568_040 |
0.0813869184 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 6 |
Hb_012055_040 |
0.0815385652 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_000479_240 |
0.0831833288 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000571_160 |
0.0866463008 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 9 |
Hb_071130_010 |
0.0868250238 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_031527_030 |
0.0870788716 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 11 |
Hb_000331_580 |
0.0879853489 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 12 |
Hb_004645_070 |
0.0883612907 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 13 |
Hb_006531_020 |
0.0890534445 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 14 |
Hb_007632_010 |
0.0891173938 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011849_040 |
0.0893997593 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_000494_050 |
0.0894235643 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_001628_070 |
0.0897633656 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_001167_050 |
0.0934238484 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 19 |
Hb_000163_270 |
0.0934297716 |
- |
- |
PREDICTED: uncharacterized protein LOC105642512 [Jatropha curcas] |
| 20 |
Hb_003581_170 |
0.0934896161 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |