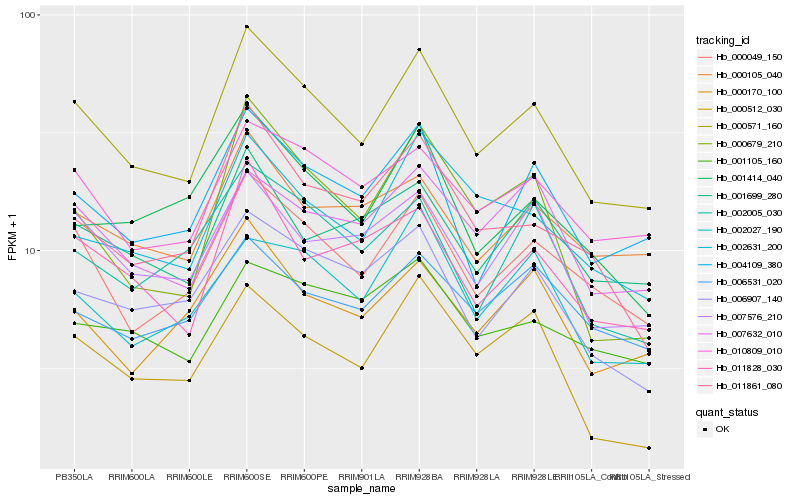
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000571_160 |
0.0 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 2 |
Hb_000049_150 |
0.0660760883 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_004109_380 |
0.0850752873 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 4 |
Hb_010809_010 |
0.0866463008 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 5 |
Hb_006907_140 |
0.0910059284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007576_210 |
0.0912978263 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 7 |
Hb_011828_030 |
0.0913611955 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 8 |
Hb_011861_080 |
0.0960626849 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 9 |
Hb_001699_280 |
0.0972650119 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 10 |
Hb_000105_040 |
0.0972717428 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 11 |
Hb_002631_200 |
0.0977727678 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000170_100 |
0.0982264507 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 13 |
Hb_002027_190 |
0.0987025444 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 14 |
Hb_007632_010 |
0.1001780942 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000679_210 |
0.1013250442 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 16 |
Hb_002005_030 |
0.101606492 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 17 |
Hb_001414_040 |
0.1036100661 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001105_160 |
0.1055218089 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 19 |
Hb_006531_020 |
0.106548016 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 20 |
Hb_000512_030 |
0.1067755411 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |