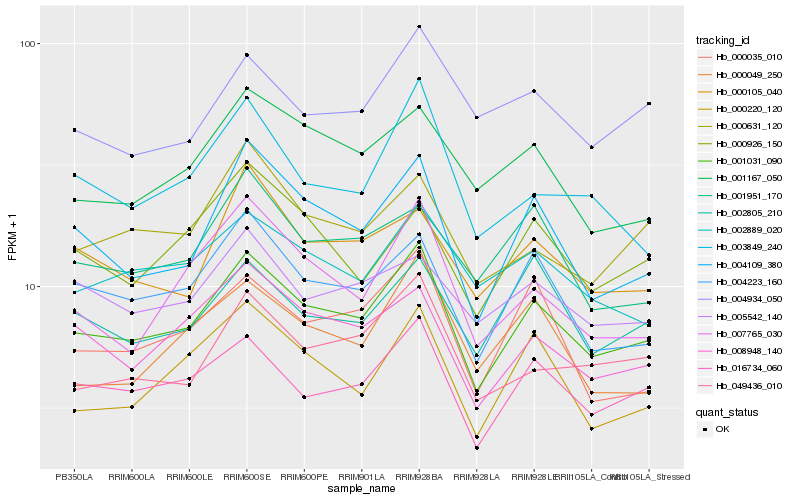
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001031_090 |
0.0 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 2 |
Hb_016734_060 |
0.0621177909 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 3 |
Hb_004109_380 |
0.0734005062 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 4 |
Hb_004223_160 |
0.0768614141 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_008948_140 |
0.080147163 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 6 |
Hb_049436_010 |
0.0853441247 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 7 |
Hb_002889_020 |
0.0879106011 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 8 |
Hb_000220_120 |
0.0891713378 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000035_010 |
0.0914395865 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001167_050 |
0.0914572425 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 11 |
Hb_003849_240 |
0.0917938261 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002805_210 |
0.091857708 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 13 |
Hb_000631_120 |
0.0918687904 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 14 |
Hb_007765_030 |
0.0930248914 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 15 |
Hb_000105_040 |
0.0942728513 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 16 |
Hb_005542_140 |
0.0957653816 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 17 |
Hb_000926_150 |
0.09589254 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_001951_170 |
0.0962910036 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 19 |
Hb_000049_250 |
0.0967921768 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004934_050 |
0.0973969413 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |