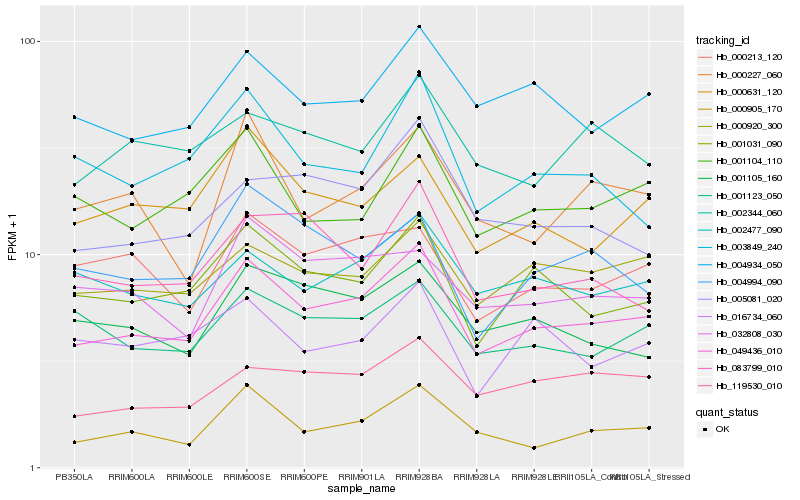
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049436_010 |
0.0 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 2 |
Hb_001031_090 |
0.0853441247 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 3 |
Hb_004934_050 |
0.0931604958 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 4 |
Hb_000905_170 |
0.0985676731 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 5 |
Hb_000920_300 |
0.0989000938 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 6 |
Hb_002477_090 |
0.0996102789 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 7 |
Hb_119530_010 |
0.1025429458 |
- |
- |
- |
| 8 |
Hb_001104_110 |
0.1033848471 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 9 |
Hb_000631_120 |
0.1034220125 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 10 |
Hb_001123_050 |
0.1037075115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001105_160 |
0.1066863131 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 12 |
Hb_005081_020 |
0.1076568014 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 13 |
Hb_000213_120 |
0.1086435232 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_002344_060 |
0.110971899 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 15 |
Hb_032808_030 |
0.1112470926 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_083799_010 |
0.11255347 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_003849_240 |
0.1128378262 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_004994_090 |
0.1151462459 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000227_060 |
0.1156720098 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 20 |
Hb_016734_060 |
0.1157661707 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |