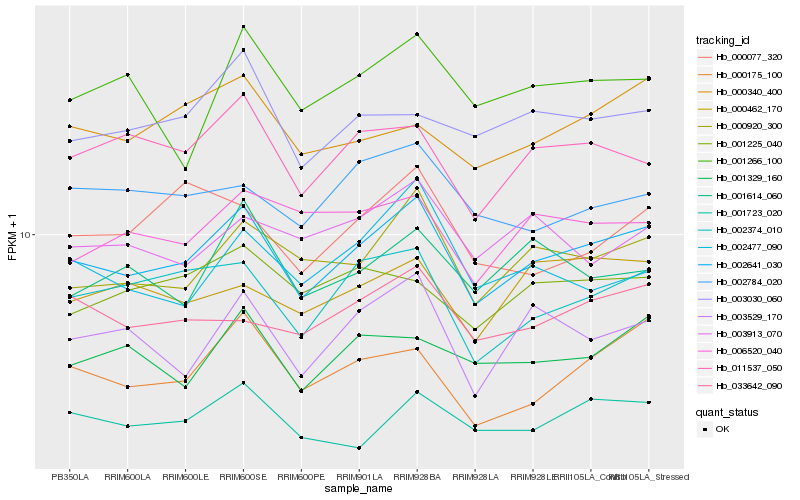
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002641_030 |
0.0 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 2 |
Hb_000920_300 |
0.0686967716 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 3 |
Hb_002374_010 |
0.0716061894 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_011537_050 |
0.0787971114 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003529_170 |
0.0795706681 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX29 [Jatropha curcas] |
| 6 |
Hb_001723_020 |
0.0805667454 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 7 |
Hb_033642_090 |
0.0807007708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000175_100 |
0.0815926681 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001329_160 |
0.085853685 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003030_060 |
0.0871726321 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 11 |
Hb_000340_400 |
0.0876562673 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 12 |
Hb_001266_100 |
0.0883759891 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 13 |
Hb_002784_020 |
0.088757673 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_000462_170 |
0.0887924155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001225_040 |
0.0894746069 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 16 |
Hb_006520_040 |
0.0896420458 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003913_070 |
0.0901565959 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 18 |
Hb_002477_090 |
0.0907402727 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001614_060 |
0.0910482558 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 20 |
Hb_000077_320 |
0.0931619047 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |