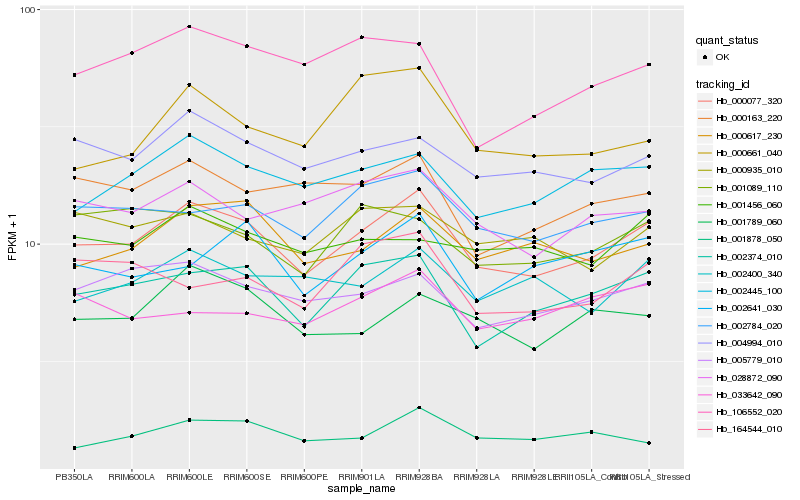
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_320 |
0.0 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 2 |
Hb_002374_010 |
0.0677453366 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002784_020 |
0.0760407556 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 4 |
Hb_004994_010 |
0.0777048774 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_002445_100 |
0.0777714508 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000617_230 |
0.0795668086 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 7 |
Hb_005779_010 |
0.0808971586 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_028872_090 |
0.0854361363 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 9 |
Hb_001789_060 |
0.0857341599 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 10 |
Hb_000163_220 |
0.0865363564 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 11 |
Hb_002400_340 |
0.0879304777 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 12 |
Hb_000935_010 |
0.0922464503 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 13 |
Hb_106552_020 |
0.0930405012 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002641_030 |
0.0931619047 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 15 |
Hb_001089_110 |
0.0933080066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001878_050 |
0.093938036 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 17 |
Hb_001456_060 |
0.0955796262 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 18 |
Hb_164544_010 |
0.0965486517 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000661_040 |
0.0970595247 |
- |
- |
PREDICTED: dihydroxy-acid dehydratase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_033642_090 |
0.0972272747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |