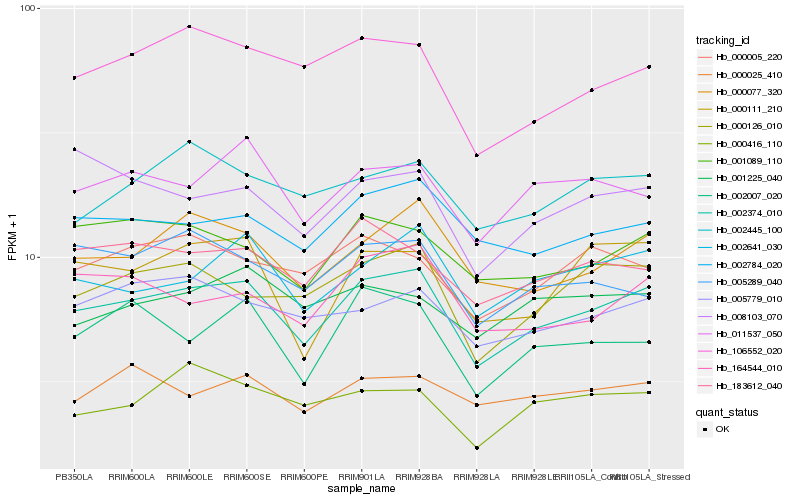
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_010 |
0.0 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000077_320 |
0.0677453366 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 3 |
Hb_002641_030 |
0.0716061894 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 4 |
Hb_000126_010 |
0.0747459037 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002784_020 |
0.0758683161 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 6 |
Hb_011537_050 |
0.077134949 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_106552_020 |
0.0781185193 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001089_110 |
0.0792302766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_183612_040 |
0.0796260035 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 10 |
Hb_164544_010 |
0.0815466599 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000111_210 |
0.0818449349 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005779_010 |
0.0822834761 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_002445_100 |
0.0824378421 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002007_020 |
0.0844437718 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like SIM [Jatropha curcas] |
| 15 |
Hb_000005_220 |
0.0850368622 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 16 |
Hb_000025_410 |
0.0861878927 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 17 |
Hb_000416_110 |
0.0872058841 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_008103_070 |
0.0887765996 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 19 |
Hb_005289_040 |
0.0890605266 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 20 |
Hb_001225_040 |
0.0893948325 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |