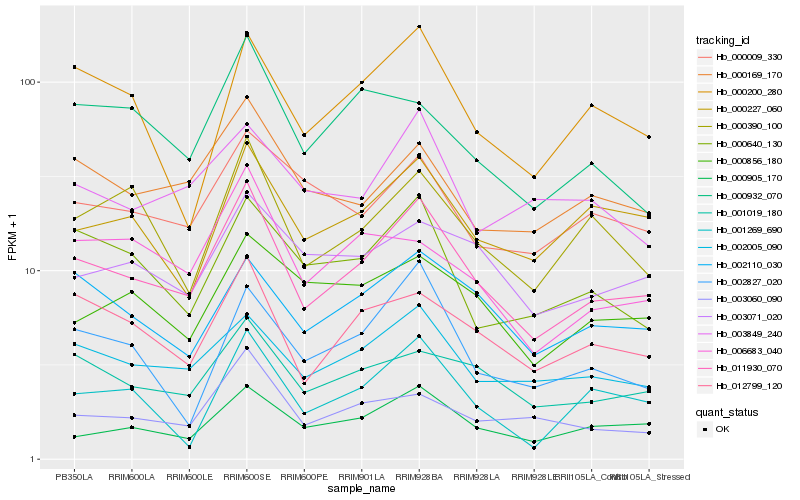
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011930_070 |
0.0 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 2 |
Hb_002005_090 |
0.1072806568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012799_120 |
0.1102218207 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001019_180 |
0.1107158327 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 5 |
Hb_000905_170 |
0.1114983237 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 6 |
Hb_002110_030 |
0.1222763459 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000227_060 |
0.1258960934 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 8 |
Hb_000169_170 |
0.1260152982 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 9 |
Hb_000390_100 |
0.1267194502 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 10 |
Hb_000200_280 |
0.1282029823 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 11 |
Hb_003060_090 |
0.1305741405 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 12 |
Hb_003071_020 |
0.1309132441 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 13 |
Hb_000640_130 |
0.1314915779 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000009_330 |
0.1369208717 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_006683_040 |
0.1379433027 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 16 |
Hb_000856_180 |
0.138848587 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 17 |
Hb_002827_020 |
0.1418885233 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 18 |
Hb_003849_240 |
0.142651332 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000932_070 |
0.1438987247 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 20 |
Hb_001269_690 |
0.1448570328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |