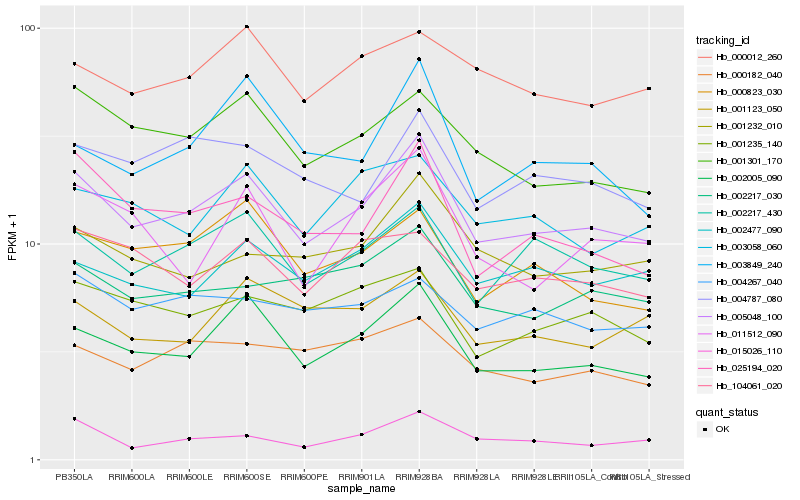
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005048_100 |
0.0 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 2 |
Hb_002005_090 |
0.0670423583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002217_430 |
0.0858483438 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 4 |
Hb_025194_020 |
0.0902550795 |
- |
- |
PREDICTED: uncharacterized protein C1F8.04c-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001301_170 |
0.090821146 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 6 |
Hb_002477_090 |
0.0916803502 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002217_030 |
0.0977687721 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001232_010 |
0.0985499839 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 9 |
Hb_000823_030 |
0.0989337131 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 10 |
Hb_004787_080 |
0.0990346354 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 11 |
Hb_015026_110 |
0.0992041809 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_003058_060 |
0.1012852425 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 13 |
Hb_001235_140 |
0.1014026046 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 14 |
Hb_000012_260 |
0.1028297069 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 15 |
Hb_104061_020 |
0.1043715396 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 16 |
Hb_001123_050 |
0.1046841228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000182_040 |
0.1048300945 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 18 |
Hb_011512_090 |
0.106706127 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 19 |
Hb_003849_240 |
0.106751753 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_004267_040 |
0.1077079158 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |