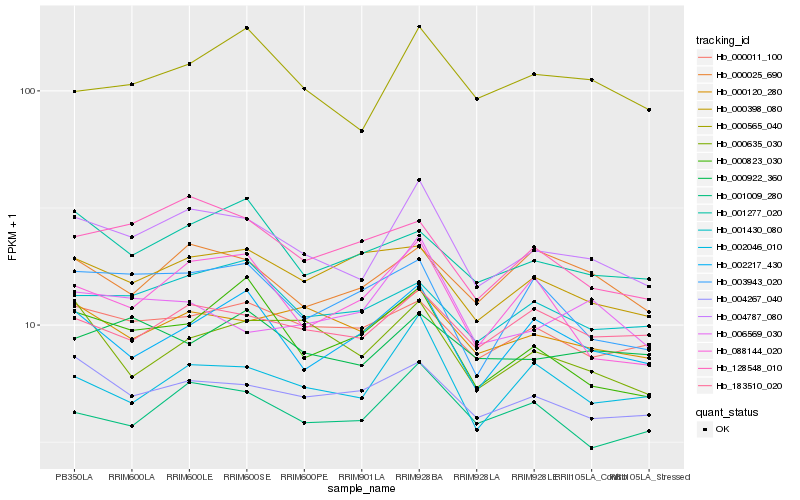
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004787_080 |
0.0 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 2 |
Hb_002046_010 |
0.0794641419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001009_280 |
0.0807387579 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 4 |
Hb_000120_280 |
0.0837751086 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 5 |
Hb_004267_040 |
0.0840792751 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 6 |
Hb_000565_040 |
0.0852391465 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 7 |
Hb_183510_020 |
0.0864242314 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 8 |
Hb_000011_100 |
0.0868083409 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 9 |
Hb_001430_080 |
0.0884064127 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 10 |
Hb_000025_690 |
0.0925701027 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 11 |
Hb_002217_430 |
0.0931657265 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 12 |
Hb_128548_010 |
0.0945803372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_088144_020 |
0.0953365854 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_006569_030 |
0.0961363916 |
- |
- |
PREDICTED: uncharacterized protein LOC105650907 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000635_030 |
0.0970132034 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 16 |
Hb_003943_020 |
0.097038046 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000922_360 |
0.0975845094 |
- |
- |
- |
| 18 |
Hb_000398_080 |
0.097963571 |
- |
- |
tip120, putative [Ricinus communis] |
| 19 |
Hb_001277_020 |
0.0980098949 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 20 |
Hb_000823_030 |
0.09892839 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |