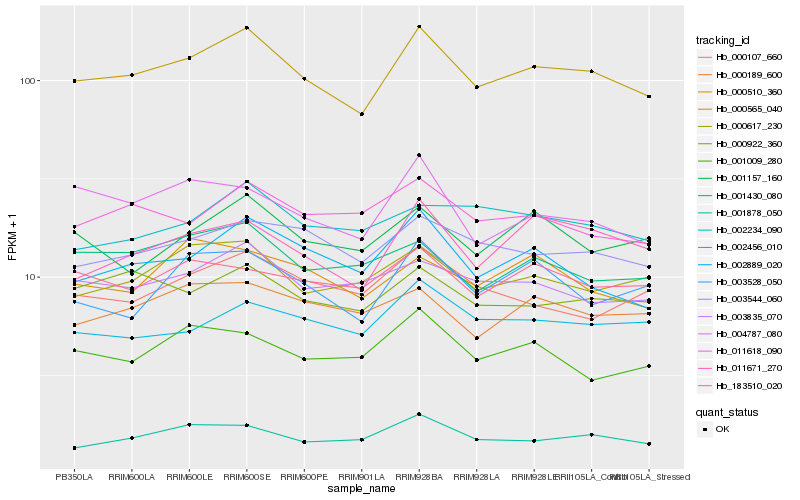
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000565_040 |
0.0 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 2 |
Hb_003544_060 |
0.0755596616 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 3 |
Hb_001878_050 |
0.0762204489 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 4 |
Hb_011671_270 |
0.0772065898 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 5 |
Hb_000617_230 |
0.0790247785 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 6 |
Hb_000922_360 |
0.0792770173 |
- |
- |
- |
| 7 |
Hb_002889_020 |
0.0793059085 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 8 |
Hb_003835_070 |
0.0804520749 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002456_010 |
0.0814106011 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_011618_090 |
0.082932078 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_003528_050 |
0.0841869407 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 12 |
Hb_004787_080 |
0.0852391465 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 13 |
Hb_000189_600 |
0.0857338081 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_001430_080 |
0.0861195644 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 15 |
Hb_183510_020 |
0.0864482607 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 16 |
Hb_001157_160 |
0.0866330027 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000107_660 |
0.0888095458 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 18 |
Hb_001009_280 |
0.0896578268 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 19 |
Hb_002234_090 |
0.0900074915 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 20 |
Hb_000510_360 |
0.0901325925 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |