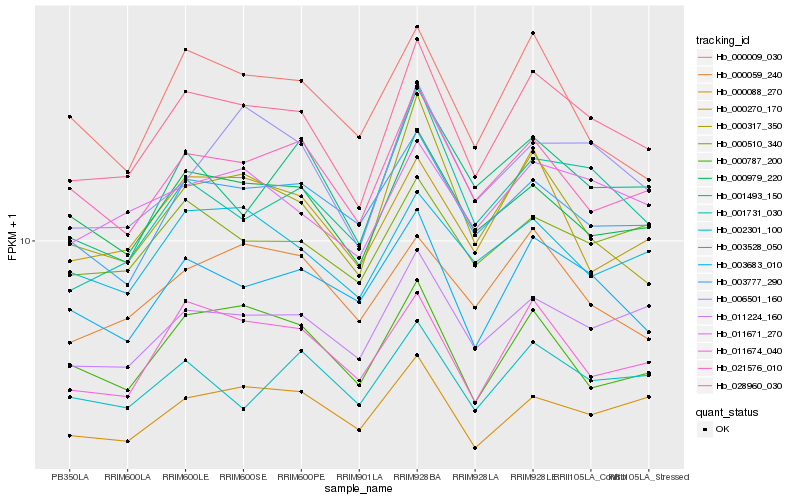
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028960_030 |
0.0 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 2 |
Hb_011224_160 |
0.0790378723 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 3 |
Hb_003683_010 |
0.0818883881 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 4 |
Hb_000979_220 |
0.0838993732 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011671_270 |
0.0847080008 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 6 |
Hb_011674_040 |
0.0861721148 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 7 |
Hb_021576_010 |
0.0869150976 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 8 |
Hb_003777_290 |
0.089389507 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 9 |
Hb_000510_340 |
0.091859403 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 10 |
Hb_000088_270 |
0.094978839 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 11 |
Hb_000317_350 |
0.0949837324 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 12 |
Hb_000270_170 |
0.0953829994 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 13 |
Hb_003528_050 |
0.0961671597 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_000787_200 |
0.0983230866 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_006501_160 |
0.0984854587 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 16 |
Hb_001493_150 |
0.0992495354 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 17 |
Hb_000059_240 |
0.0999289781 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 18 |
Hb_001731_030 |
0.1009501483 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 19 |
Hb_002301_100 |
0.1024140253 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 20 |
Hb_000009_030 |
0.1026895894 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |