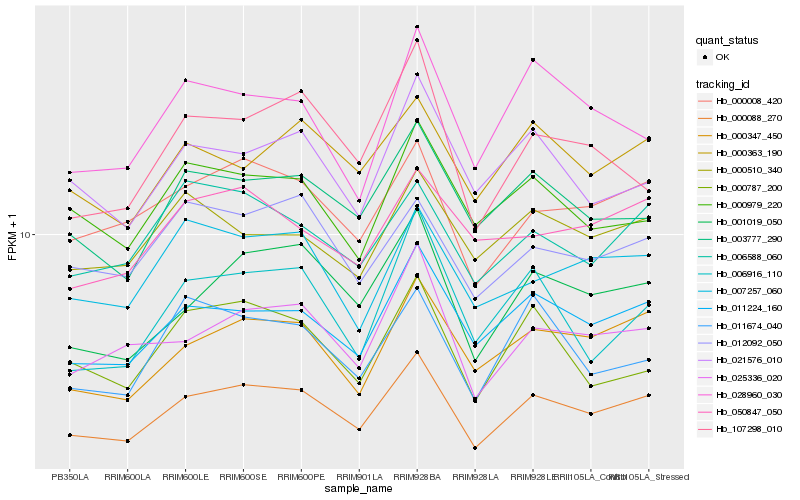
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 2 |
Hb_001019_050 |
0.0661382047 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 3 |
Hb_011224_160 |
0.0674463203 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 4 |
Hb_000347_450 |
0.0690977585 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000008_420 |
0.0725693238 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 6 |
Hb_025336_020 |
0.0779537102 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 7 |
Hb_003777_290 |
0.0833280228 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 8 |
Hb_000363_190 |
0.0912807796 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_007257_060 |
0.0922700741 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 10 |
Hb_006588_060 |
0.0938753939 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 11 |
Hb_021576_010 |
0.0949676693 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 12 |
Hb_028960_030 |
0.094978839 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 13 |
Hb_012092_050 |
0.0955286447 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_107298_010 |
0.0956605977 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 15 |
Hb_006916_110 |
0.0958244669 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000979_220 |
0.098132714 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000787_200 |
0.0992209672 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_011674_040 |
0.0999481652 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 19 |
Hb_050847_050 |
0.0999908614 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000510_340 |
0.1001580574 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |