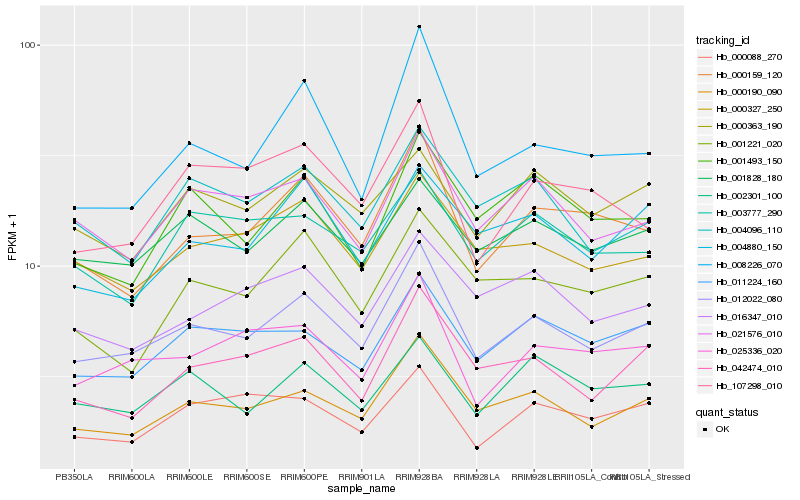
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012022_080 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000190_090 |
0.0561232652 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 3 |
Hb_000159_120 |
0.0759697711 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 4 |
Hb_025336_020 |
0.0818342834 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 5 |
Hb_021576_010 |
0.0828805426 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 6 |
Hb_000327_250 |
0.0831739688 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 7 |
Hb_011224_160 |
0.0848304566 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 8 |
Hb_001828_180 |
0.0857796374 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 9 |
Hb_042474_010 |
0.0876649993 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 10 |
Hb_008226_070 |
0.0877421452 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 11 |
Hb_004096_110 |
0.0905701138 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 12 |
Hb_016347_010 |
0.0937911759 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_002301_100 |
0.0951558018 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 14 |
Hb_004880_150 |
0.0964030355 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 15 |
Hb_001493_150 |
0.097288233 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 16 |
Hb_000363_190 |
0.0989281084 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003777_290 |
0.0992499369 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 18 |
Hb_107298_010 |
0.1009289699 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 19 |
Hb_000088_270 |
0.1010789262 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 20 |
Hb_001221_020 |
0.1011116946 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |