| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
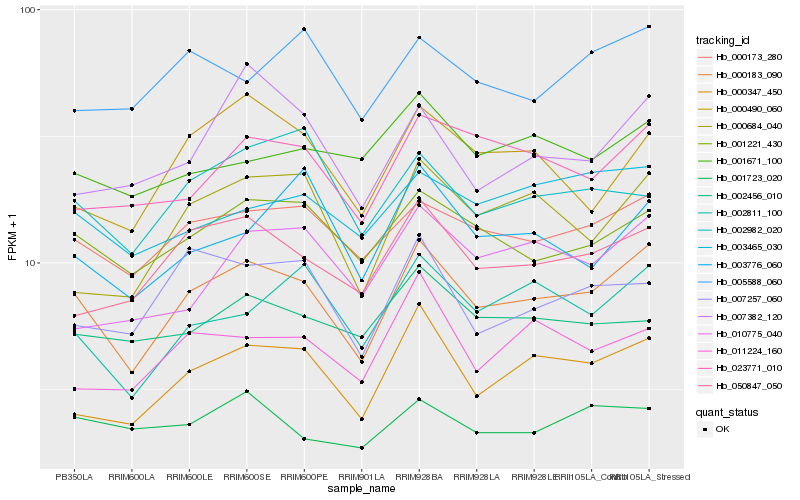
Hb_000183_090 |
0.0 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000173_280 |
0.0832382005 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 3 |
Hb_000347_450 |
0.0873509088 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_001221_430 |
0.0912247819 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At5g63520 isoform X2 [Jatropha curcas] |
| 5 |
Hb_050847_050 |
0.0977925397 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002811_100 |
0.1003106442 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 7 |
Hb_023771_010 |
0.1027660844 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000684_040 |
0.1042027408 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 9 |
Hb_003465_030 |
0.1059460503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000490_060 |
0.1078679644 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 11 |
Hb_007257_060 |
0.1116173062 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 12 |
Hb_005588_060 |
0.1132732604 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 13 |
Hb_007382_120 |
0.1138234411 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_011224_160 |
0.1139027537 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 15 |
Hb_003776_060 |
0.1153134257 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 16 |
Hb_010775_040 |
0.1153856066 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 17 |
Hb_001671_100 |
0.1157932858 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 18 |
Hb_001723_020 |
0.1159998884 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 19 |
Hb_002982_020 |
0.1166982752 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 20 |
Hb_002456_010 |
0.1167656028 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |