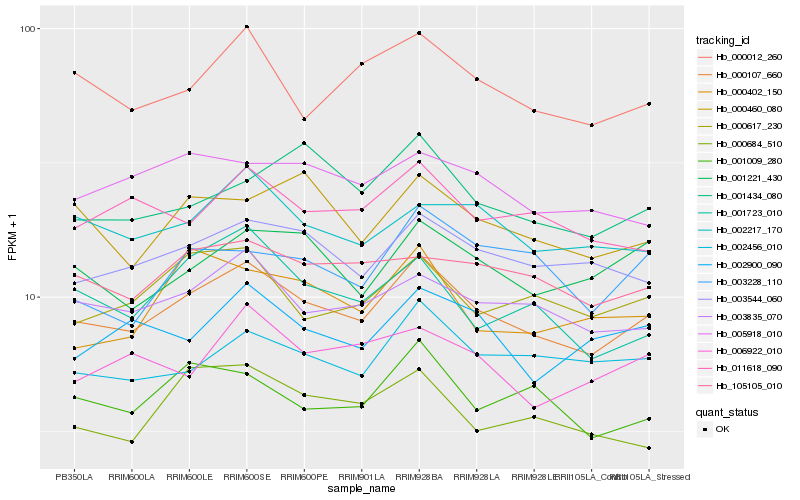
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_660 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 2 |
Hb_003835_070 |
0.0651921019 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003544_060 |
0.0675689017 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 4 |
Hb_000012_260 |
0.0731637756 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 5 |
Hb_002456_010 |
0.0740513965 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_105105_010 |
0.074500708 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 7 |
Hb_002217_170 |
0.0749495378 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 8 |
Hb_000617_230 |
0.0759373093 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 9 |
Hb_001009_280 |
0.0767320434 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 10 |
Hb_001221_430 |
0.0770736197 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At5g63520 isoform X2 [Jatropha curcas] |
| 11 |
Hb_011618_090 |
0.077431353 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_001723_010 |
0.0803181864 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 13 |
Hb_002900_090 |
0.0808389922 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 14 |
Hb_005918_010 |
0.0820100234 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 15 |
Hb_000460_080 |
0.082238837 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 16 |
Hb_006922_010 |
0.0834566732 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 17 |
Hb_001434_080 |
0.0834934049 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 18 |
Hb_000684_510 |
0.0835817302 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 19 |
Hb_003228_110 |
0.0853754718 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 20 |
Hb_000402_150 |
0.0854711279 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |