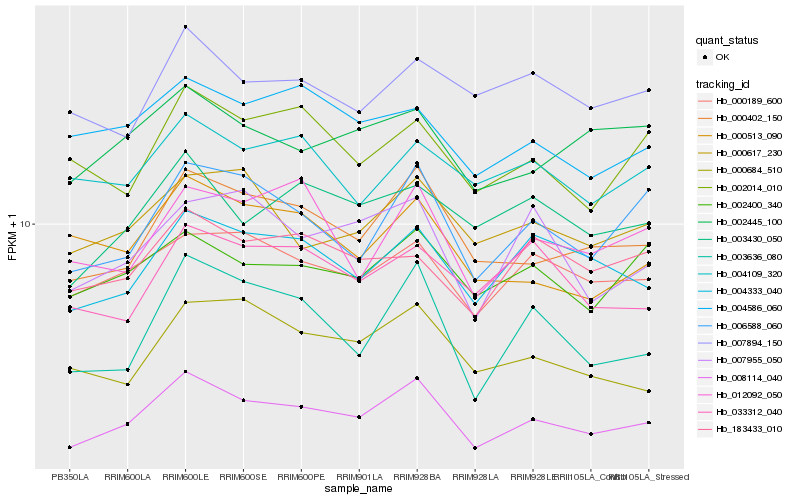
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008114_040 |
0.0 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 2 |
Hb_000402_150 |
0.0525333648 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000189_600 |
0.0679026004 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_004333_040 |
0.0696551351 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 5 |
Hb_006588_060 |
0.071450576 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 6 |
Hb_002400_340 |
0.0743344206 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 7 |
Hb_000617_230 |
0.0758358378 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 8 |
Hb_003430_050 |
0.0760541169 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 9 |
Hb_002445_100 |
0.0786327791 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000684_510 |
0.0801249331 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 11 |
Hb_004586_060 |
0.0808738817 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 12 |
Hb_183433_010 |
0.082732992 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 13 |
Hb_012092_050 |
0.0835769417 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003636_080 |
0.0836901017 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 15 |
Hb_002014_010 |
0.0838720911 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_007955_050 |
0.0840388507 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_000513_090 |
0.0841896363 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 18 |
Hb_004109_320 |
0.0843178847 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 19 |
Hb_007894_150 |
0.0852616587 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 20 |
Hb_033312_040 |
0.0853844602 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |