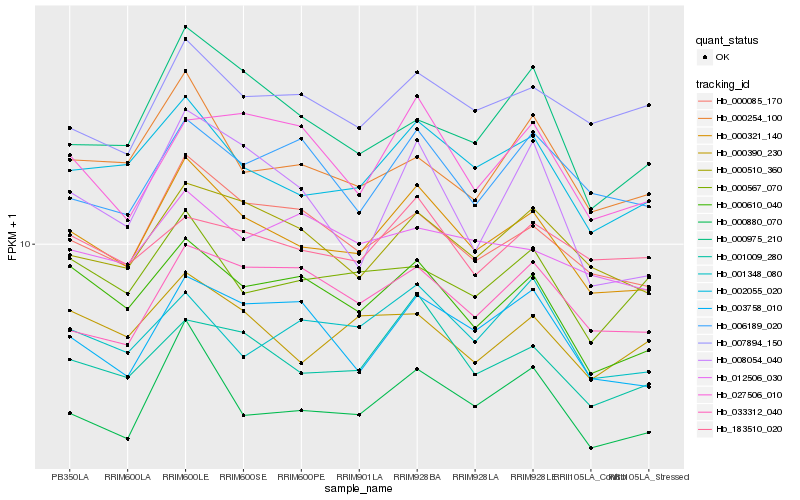
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000321_140 |
0.0 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 2 |
Hb_002055_020 |
0.0518832582 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 3 |
Hb_000085_170 |
0.0595136938 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001009_280 |
0.0601812364 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 5 |
Hb_000610_040 |
0.0633556155 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000510_360 |
0.0638056453 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 7 |
Hb_000880_070 |
0.0647355285 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 8 |
Hb_003758_010 |
0.0724202828 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000254_100 |
0.0791471568 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 10 |
Hb_033312_040 |
0.080487896 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007894_150 |
0.0805684627 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 12 |
Hb_000390_230 |
0.080609408 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 13 |
Hb_006189_020 |
0.0836026713 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 14 |
Hb_027506_010 |
0.0837694433 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 15 |
Hb_008054_040 |
0.084643724 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 16 |
Hb_000975_210 |
0.0846623555 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 17 |
Hb_183510_020 |
0.0851577238 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 18 |
Hb_000567_070 |
0.0853459411 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 19 |
Hb_001348_080 |
0.0853838394 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 20 |
Hb_012506_030 |
0.0855110428 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |