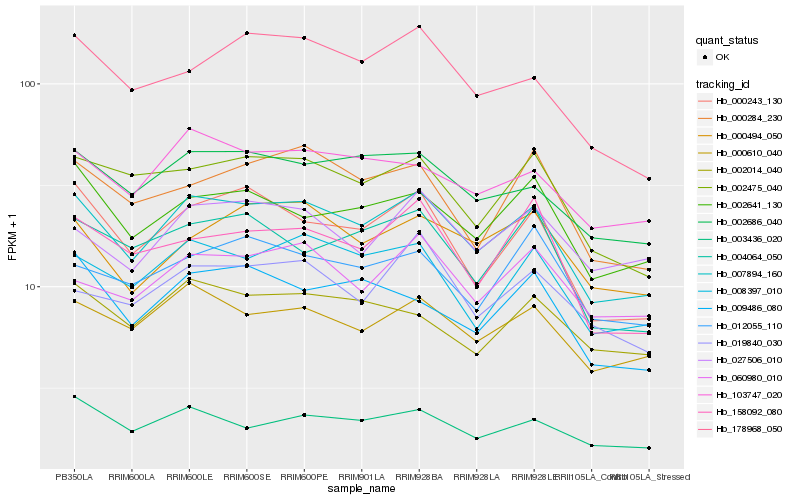
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_160 |
0.0 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 2 |
Hb_000610_040 |
0.0687656555 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008397_010 |
0.0693215907 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002686_040 |
0.071274874 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 5 |
Hb_000243_130 |
0.0724425772 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 6 |
Hb_103747_020 |
0.0773006139 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 7 |
Hb_002475_040 |
0.077308978 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002641_130 |
0.0785455285 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 9 |
Hb_003436_020 |
0.0792246811 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 10 |
Hb_178968_050 |
0.082256714 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 11 |
Hb_000494_050 |
0.0834572488 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_000284_230 |
0.0843134467 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 13 |
Hb_158092_080 |
0.0849156518 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 14 |
Hb_004064_050 |
0.085729465 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 15 |
Hb_060980_010 |
0.0858945768 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_009486_080 |
0.0882424037 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 17 |
Hb_002014_040 |
0.0884467158 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 18 |
Hb_027506_010 |
0.0887657646 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 19 |
Hb_019840_030 |
0.0897393525 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 20 |
Hb_012055_110 |
0.0899970503 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |