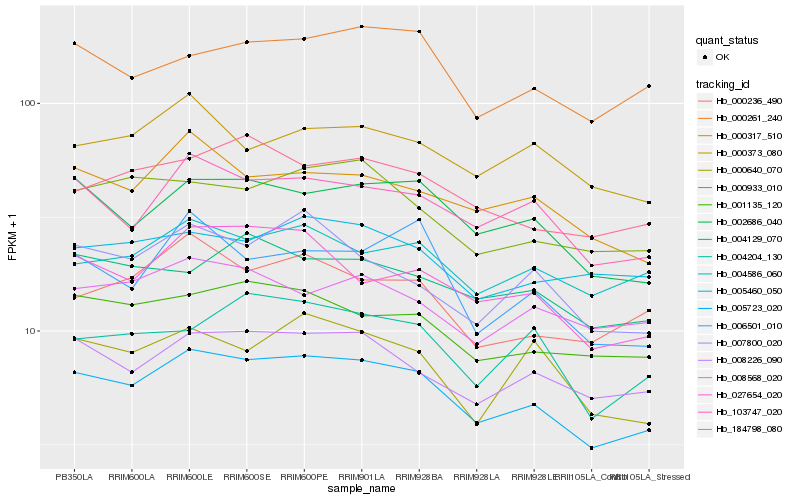
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005723_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002686_040 |
0.061402793 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 3 |
Hb_103747_020 |
0.0629308177 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 4 |
Hb_008226_090 |
0.0642259995 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001135_120 |
0.0704220656 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_027654_020 |
0.0704870615 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 7 |
Hb_000933_010 |
0.0732482133 |
- |
- |
- |
| 8 |
Hb_000317_510 |
0.0740528895 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 9 |
Hb_184798_080 |
0.0748039459 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 10 |
Hb_000261_240 |
0.0754201961 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 11 |
Hb_007800_020 |
0.0769844128 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000236_490 |
0.0775727562 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 13 |
Hb_004204_130 |
0.0778120198 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 14 |
Hb_004586_060 |
0.0787451559 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_000640_070 |
0.0808540232 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 16 |
Hb_004129_070 |
0.0813402578 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 17 |
Hb_008568_020 |
0.0824215955 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 18 |
Hb_006501_010 |
0.083458245 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 19 |
Hb_005460_050 |
0.0835793102 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 20 |
Hb_000373_080 |
0.0837130971 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |