| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
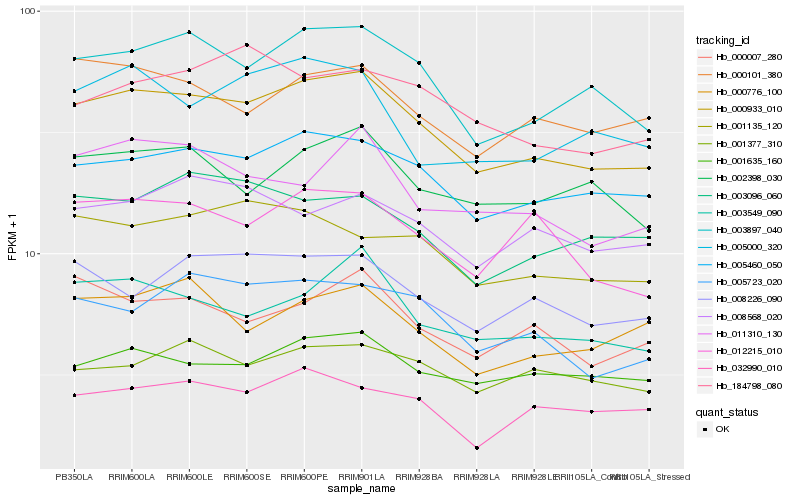
Hb_000933_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_005460_050 |
0.0540930473 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 3 |
Hb_003897_040 |
0.055352259 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 4 |
Hb_003549_090 |
0.0686750307 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 5 |
Hb_005723_020 |
0.0732482133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002398_030 |
0.0739222846 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_008226_090 |
0.0751073345 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000101_380 |
0.0756299231 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 9 |
Hb_001635_160 |
0.0756350805 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 10 |
Hb_012215_010 |
0.0759630338 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 11 |
Hb_000007_280 |
0.076305771 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 12 |
Hb_001135_120 |
0.0778428355 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_011310_130 |
0.0790406275 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000776_100 |
0.0811828305 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 15 |
Hb_008568_020 |
0.0818162407 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 16 |
Hb_005000_320 |
0.081952694 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 17 |
Hb_001377_310 |
0.0819960552 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 18 |
Hb_032990_010 |
0.0824191405 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 19 |
Hb_003096_060 |
0.0825802245 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 20 |
Hb_184798_080 |
0.08291051 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |