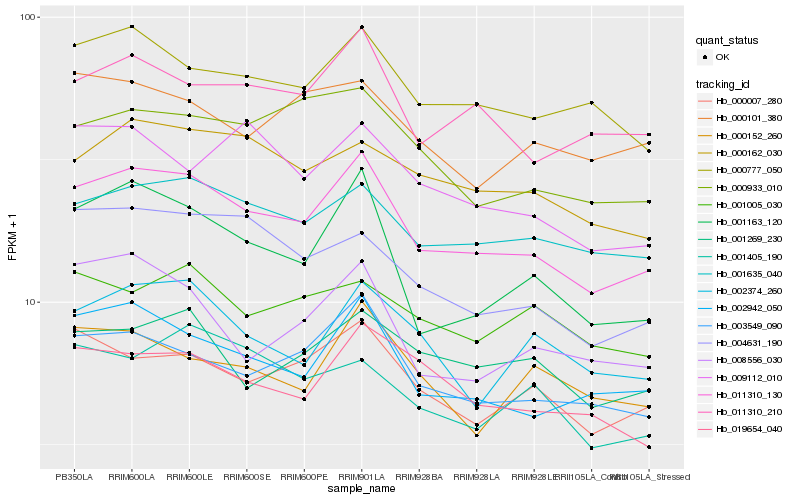
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_130 |
0.0 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_002942_050 |
0.0588781864 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003549_090 |
0.0611532034 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 4 |
Hb_001635_040 |
0.0669599854 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 5 |
Hb_000777_050 |
0.0675031075 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 6 |
Hb_001163_120 |
0.0688148171 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_011310_210 |
0.069854146 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 8 |
Hb_000007_280 |
0.0713072403 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 9 |
Hb_000162_030 |
0.0758335176 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_004631_190 |
0.0767144397 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 11 |
Hb_019654_040 |
0.0787186958 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000933_010 |
0.0790406275 |
- |
- |
- |
| 13 |
Hb_001269_230 |
0.0808405416 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 14 |
Hb_002374_260 |
0.0808580195 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 15 |
Hb_009112_010 |
0.0836022681 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 16 |
Hb_008556_030 |
0.0838253264 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000101_380 |
0.0874660744 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000152_260 |
0.0875584498 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 19 |
Hb_001005_030 |
0.0875821587 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_001405_190 |
0.0880596973 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |