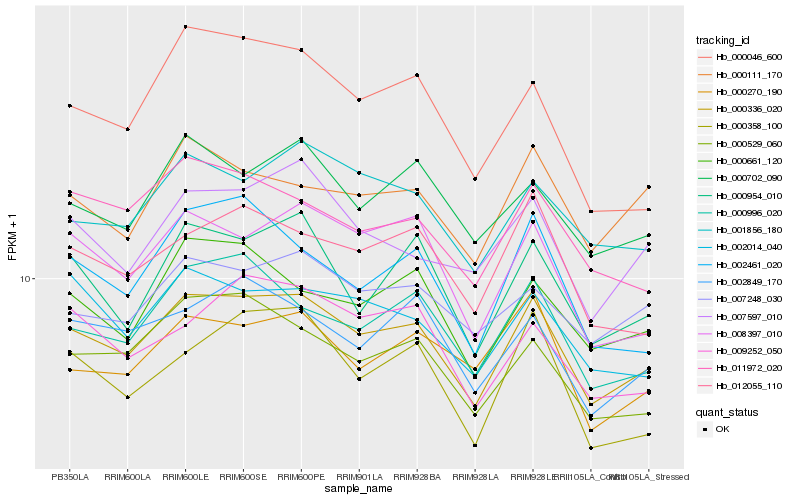
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_020 |
0.0 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 2 |
Hb_000996_020 |
0.0658272055 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 3 |
Hb_008397_010 |
0.0660649789 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000529_060 |
0.0670086392 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001856_180 |
0.0677926436 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 6 |
Hb_012055_110 |
0.0681295876 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 7 |
Hb_002849_170 |
0.0687234621 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000702_090 |
0.0688338993 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 9 |
Hb_000661_120 |
0.0690443742 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 10 |
Hb_000954_010 |
0.0697545315 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 11 |
Hb_002014_040 |
0.0698459053 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 12 |
Hb_007248_030 |
0.0703367564 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 13 |
Hb_011972_020 |
0.0709755066 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_007597_010 |
0.0723033508 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 15 |
Hb_000111_170 |
0.0757225285 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009252_050 |
0.0758341886 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_002461_020 |
0.0763564383 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000270_190 |
0.076480939 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 19 |
Hb_000046_600 |
0.0785336455 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 20 |
Hb_000358_100 |
0.0787691753 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |