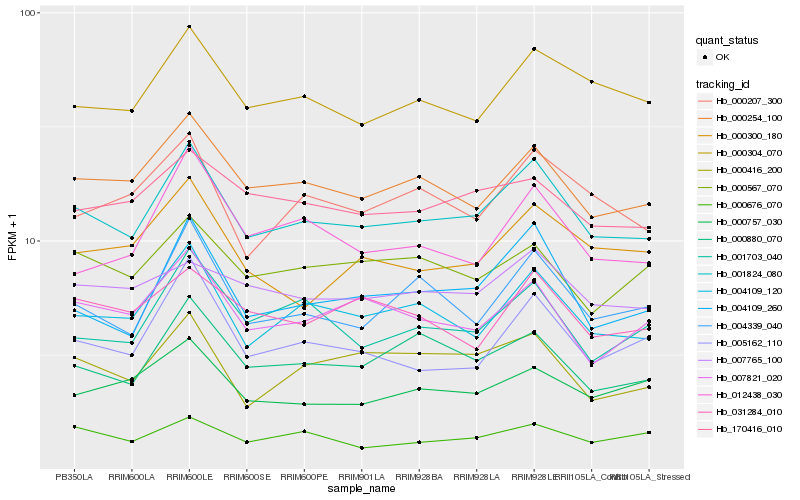
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001824_080 |
0.0 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 2 |
Hb_004109_120 |
0.0629988904 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000254_100 |
0.0649248395 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 4 |
Hb_004109_260 |
0.0679689506 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 5 |
Hb_000304_070 |
0.0730175452 |
- |
- |
non-canonical ubiquitin conjugating enzyme, putative [Ricinus communis] |
| 6 |
Hb_004339_040 |
0.0778625589 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 7 |
Hb_007821_020 |
0.0806837965 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000880_070 |
0.0807150294 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 9 |
Hb_000207_300 |
0.0835926335 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001703_040 |
0.0858344032 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000416_200 |
0.0868551733 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 12 |
Hb_170416_010 |
0.0868698974 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 13 |
Hb_005162_110 |
0.0870939814 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 14 |
Hb_000567_070 |
0.0893446442 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 15 |
Hb_007765_100 |
0.0894754628 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 16 |
Hb_000300_180 |
0.0900735347 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 17 |
Hb_012438_030 |
0.0906496023 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 18 |
Hb_000676_070 |
0.0911927109 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 19 |
Hb_000757_030 |
0.0916380423 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 20 |
Hb_031284_010 |
0.0923959836 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |