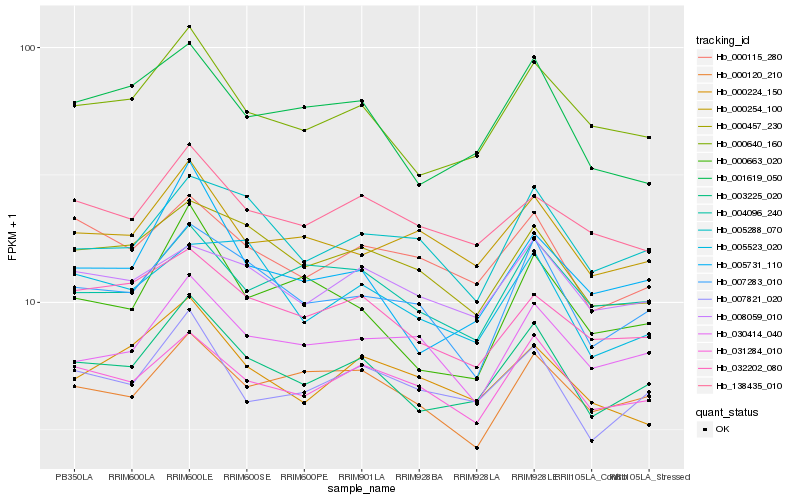
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003225_020 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 2 |
Hb_000640_160 |
0.0517665656 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 3 |
Hb_007821_020 |
0.0677933503 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 4 |
Hb_031284_010 |
0.0717718461 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 5 |
Hb_001619_050 |
0.072844718 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007283_010 |
0.0739752289 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 7 |
Hb_005731_110 |
0.0753481733 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 8 |
Hb_000115_280 |
0.0771117319 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_000457_230 |
0.0783673153 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_032202_080 |
0.079566057 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004096_240 |
0.0811770896 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 12 |
Hb_138435_010 |
0.0815049483 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 13 |
Hb_000120_210 |
0.0832559531 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 14 |
Hb_000254_100 |
0.0834314809 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 15 |
Hb_008059_010 |
0.0836831618 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005288_070 |
0.0862061067 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000663_020 |
0.08728206 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_000224_150 |
0.0884715175 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_030414_040 |
0.0888007276 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005523_020 |
0.0894155333 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |