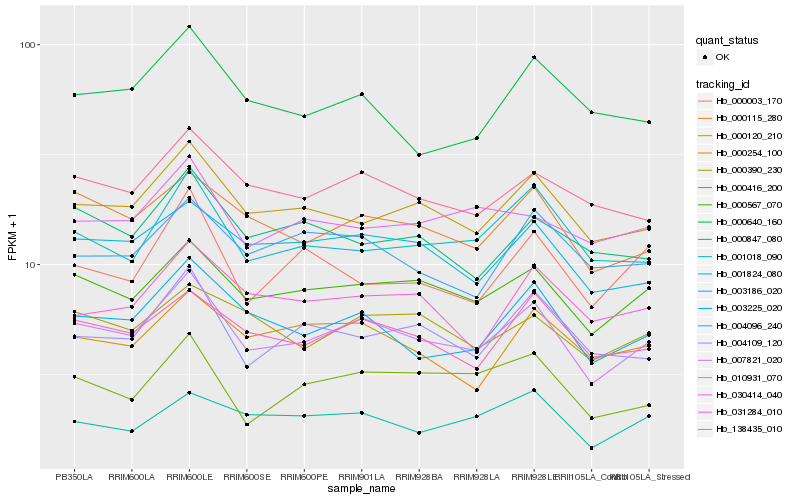
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007821_020 |
0.0 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000567_070 |
0.0591766282 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 3 |
Hb_000254_100 |
0.0671765655 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 4 |
Hb_003225_020 |
0.0677933503 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 5 |
Hb_000115_280 |
0.0695623447 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 6 |
Hb_031284_010 |
0.0717917643 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 7 |
Hb_003186_020 |
0.07580212 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 8 |
Hb_138435_010 |
0.0788556681 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 9 |
Hb_001824_080 |
0.0806837965 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 10 |
Hb_004109_120 |
0.0824852415 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000416_200 |
0.0889987612 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 12 |
Hb_000390_230 |
0.0890447155 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 13 |
Hb_030414_040 |
0.0892815484 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004096_240 |
0.0893519365 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 15 |
Hb_000120_210 |
0.08970019 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 16 |
Hb_001018_090 |
0.0912726369 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 17 |
Hb_000847_080 |
0.0912885631 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 18 |
Hb_000640_160 |
0.0916686079 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 19 |
Hb_000003_170 |
0.0935450567 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 20 |
Hb_010931_070 |
0.0937096192 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |