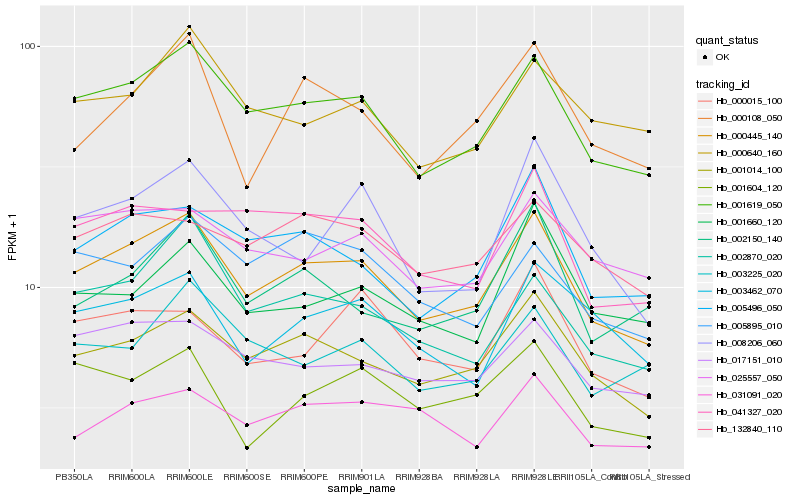
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_140 |
0.0 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 2 |
Hb_001619_050 |
0.0399310517 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001014_100 |
0.0613544854 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 4 |
Hb_005496_050 |
0.0805877212 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 5 |
Hb_017151_010 |
0.0856365669 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 6 |
Hb_025557_050 |
0.0893617269 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 7 |
Hb_002870_020 |
0.0897051071 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000108_050 |
0.0908135624 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 9 |
Hb_132840_110 |
0.092007401 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_041327_020 |
0.0930987016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003462_070 |
0.0942610046 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 12 |
Hb_000640_160 |
0.0945064594 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 13 |
Hb_002150_140 |
0.0955837975 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 14 |
Hb_005895_010 |
0.095772894 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001604_120 |
0.0959976947 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000015_100 |
0.0977814729 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_031091_020 |
0.0979178683 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 18 |
Hb_003225_020 |
0.098241109 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 19 |
Hb_008206_060 |
0.0984018844 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001660_120 |
0.0986650971 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |