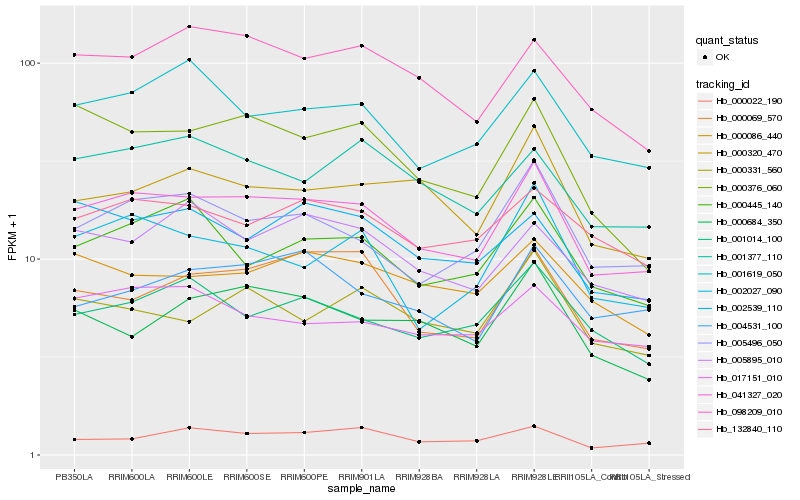
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041327_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005496_050 |
0.0799137661 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 3 |
Hb_098209_010 |
0.0894212949 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 4 |
Hb_132840_110 |
0.0897043075 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_001619_050 |
0.0898240796 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000320_470 |
0.0922627519 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 7 |
Hb_000445_140 |
0.0930987016 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 8 |
Hb_001014_100 |
0.0931563207 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 9 |
Hb_000376_060 |
0.0944908951 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001377_110 |
0.095370899 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 11 |
Hb_000069_570 |
0.0961070994 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000086_440 |
0.0961402916 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 13 |
Hb_002027_090 |
0.0969588818 |
- |
- |
- |
| 14 |
Hb_017151_010 |
0.0970858203 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 15 |
Hb_000331_560 |
0.0972814626 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 16 |
Hb_005895_010 |
0.0978830855 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004531_100 |
0.0985130318 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000022_190 |
0.0993929066 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 19 |
Hb_000684_350 |
0.0995598956 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002539_110 |
0.099610841 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |