| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
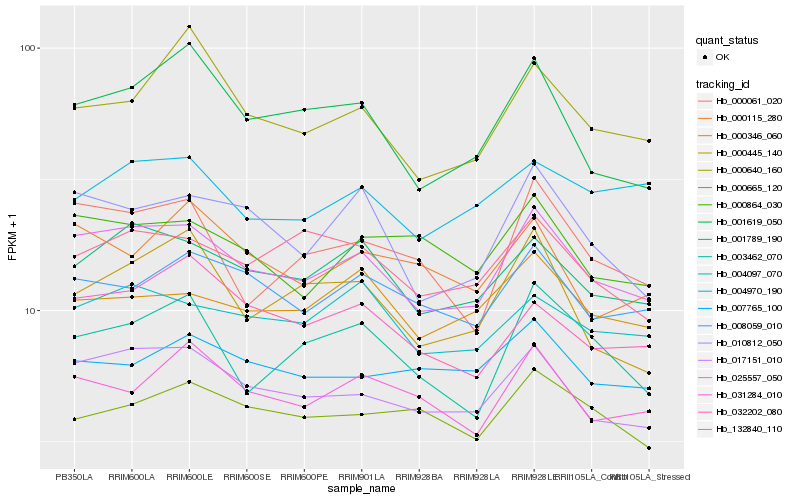
Hb_025557_050 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 2 |
Hb_017151_010 |
0.0473575065 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 3 |
Hb_001789_190 |
0.0573874558 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 4 |
Hb_010812_050 |
0.0696091871 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008059_010 |
0.072293991 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004097_070 |
0.074581445 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 7 |
Hb_031284_010 |
0.077325362 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 8 |
Hb_132840_110 |
0.0776963189 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_001619_050 |
0.0780297293 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 10 |
Hb_032202_080 |
0.0787897408 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000864_030 |
0.0806403757 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000346_060 |
0.0811987582 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 13 |
Hb_000061_020 |
0.0826451312 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 14 |
Hb_000640_160 |
0.0840255732 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 15 |
Hb_003462_070 |
0.0858910845 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 16 |
Hb_004970_190 |
0.0877018432 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000115_280 |
0.0877646976 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000665_120 |
0.088919555 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 19 |
Hb_007765_100 |
0.0889296534 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 20 |
Hb_000445_140 |
0.0893617269 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |