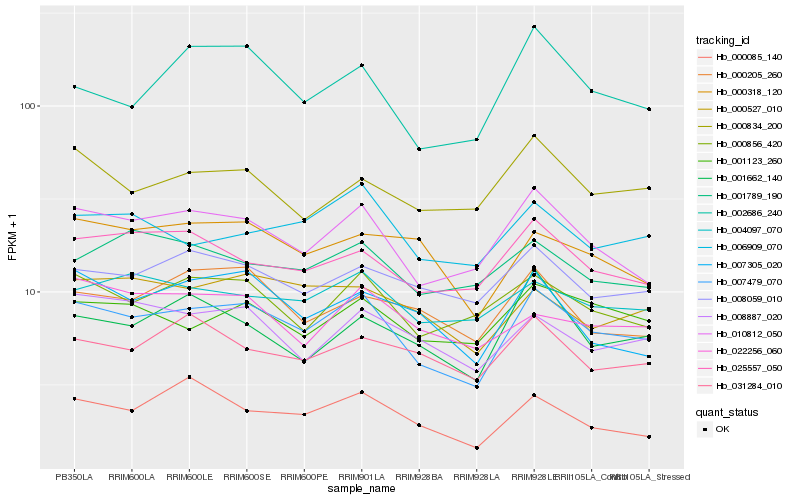
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007479_070 |
0.0 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 2 |
Hb_010812_050 |
0.0715265921 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000856_420 |
0.0908677145 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_025557_050 |
0.0959517704 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 5 |
Hb_001123_260 |
0.0980580794 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_002686_240 |
0.098325498 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 7 |
Hb_008059_010 |
0.099294209 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004097_070 |
0.099374708 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 9 |
Hb_000527_010 |
0.1003539497 |
- |
- |
katanin P80 subunit, putative [Ricinus communis] |
| 10 |
Hb_006909_070 |
0.1018277758 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_008887_020 |
0.1024855599 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 12 |
Hb_000085_140 |
0.102544445 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 13 |
Hb_007305_020 |
0.1034833345 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 14 |
Hb_000834_200 |
0.1044388448 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 15 |
Hb_000318_120 |
0.1056738497 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001662_140 |
0.1068767109 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000205_260 |
0.1073233275 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 18 |
Hb_001789_190 |
0.1077743021 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 19 |
Hb_031284_010 |
0.1079294872 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 20 |
Hb_022256_060 |
0.1079737545 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |