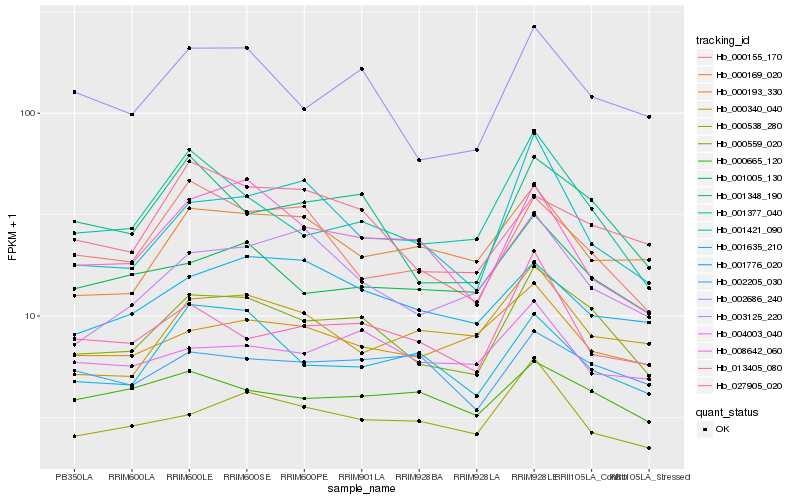
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000559_020 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 2 |
Hb_002686_240 |
0.0831016383 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 3 |
Hb_001348_190 |
0.08489961 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001377_040 |
0.0994171909 |
- |
- |
chitinase, putative [Ricinus communis] |
| 5 |
Hb_001005_130 |
0.1004672037 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000538_280 |
0.1059598647 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 7 |
Hb_027905_020 |
0.1102860002 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 8 |
Hb_001635_210 |
0.1121511634 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003125_220 |
0.1125898265 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 10 |
Hb_000193_330 |
0.1144233134 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 11 |
Hb_001776_020 |
0.1151983326 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 12 |
Hb_013405_080 |
0.1152870419 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 13 |
Hb_000665_120 |
0.1154623525 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 14 |
Hb_000169_020 |
0.1155552006 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 15 |
Hb_002205_030 |
0.1157039059 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 16 |
Hb_008642_060 |
0.1181527108 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 17 |
Hb_000155_170 |
0.1181705903 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 18 |
Hb_001421_090 |
0.1195475884 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 19 |
Hb_004003_040 |
0.1196930295 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000340_040 |
0.1199945689 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |