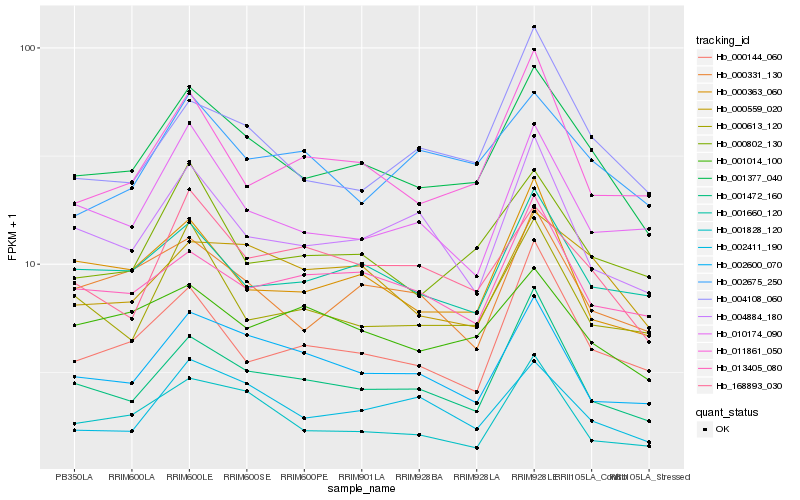
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_040 |
0.0 |
- |
- |
chitinase, putative [Ricinus communis] |
| 2 |
Hb_000802_130 |
0.0955933873 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 3 |
Hb_001472_160 |
0.0970914862 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 4 |
Hb_001660_120 |
0.0971913168 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000559_020 |
0.0994171909 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 6 |
Hb_000331_130 |
0.1019638323 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002411_190 |
0.1038001937 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 8 |
Hb_168893_030 |
0.104584979 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 9 |
Hb_002600_070 |
0.1047620393 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 10 |
Hb_000613_120 |
0.1058400221 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000363_060 |
0.1063739143 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004108_060 |
0.1067052189 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 13 |
Hb_001828_120 |
0.1070393291 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 14 |
Hb_010174_090 |
0.1081097264 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004884_180 |
0.1087615821 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 16 |
Hb_013405_080 |
0.1096948842 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_000144_060 |
0.1100800887 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_011861_050 |
0.1110681977 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_001014_100 |
0.1111803642 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 20 |
Hb_002675_250 |
0.1114906588 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |