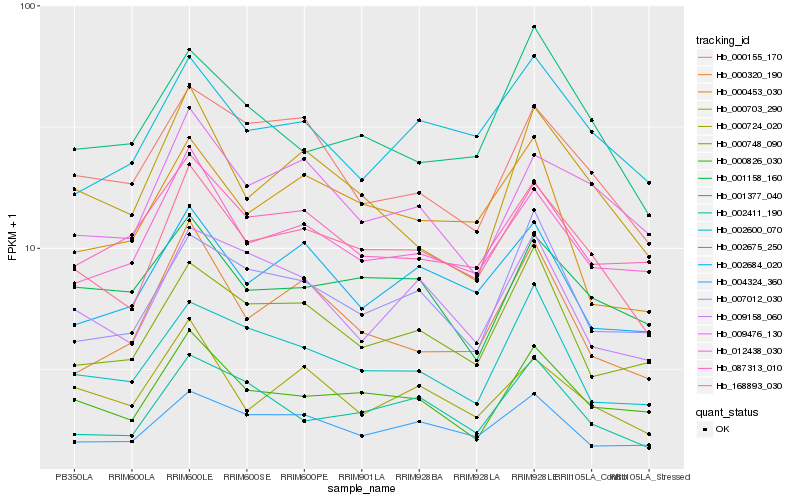
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168893_030 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 2 |
Hb_000826_030 |
0.090198342 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 3 |
Hb_012438_030 |
0.0906664134 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 4 |
Hb_004324_360 |
0.0911542063 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002411_190 |
0.0919539175 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 6 |
Hb_002675_250 |
0.0961099967 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_002684_020 |
0.096226406 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 8 |
Hb_009476_130 |
0.0974092514 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 9 |
Hb_000724_020 |
0.0982011464 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 10 |
Hb_000155_170 |
0.1025738591 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 11 |
Hb_001377_040 |
0.104584979 |
- |
- |
chitinase, putative [Ricinus communis] |
| 12 |
Hb_000748_090 |
0.1057087034 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 13 |
Hb_007012_030 |
0.1059027761 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001158_160 |
0.1068212435 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002600_070 |
0.1069758659 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 16 |
Hb_000453_030 |
0.1077932384 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 17 |
Hb_087313_010 |
0.1080648607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000320_190 |
0.1082562035 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_009158_060 |
0.1084901918 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 20 |
Hb_000703_290 |
0.1089593266 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |