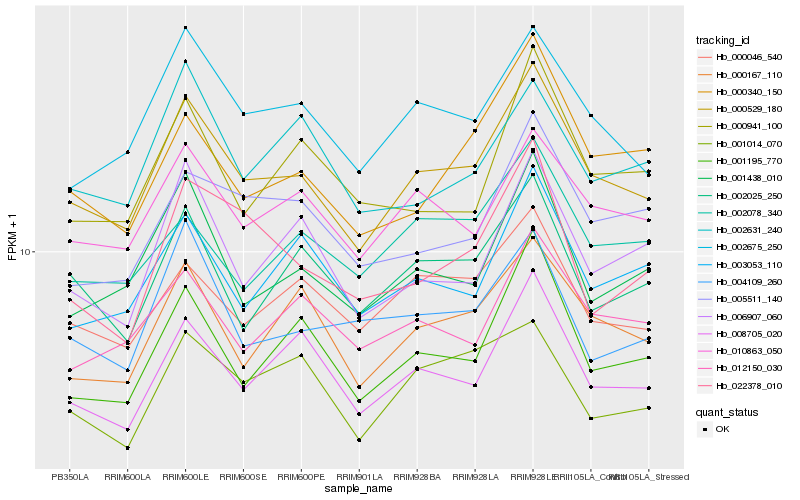
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_180 |
0.0 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002675_250 |
0.0694543388 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_010863_050 |
0.0719822048 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 4 |
Hb_008705_020 |
0.0789137188 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_000167_110 |
0.0794961981 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001438_010 |
0.0823283893 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000046_540 |
0.0826234917 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 8 |
Hb_001195_770 |
0.087097647 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 9 |
Hb_002025_250 |
0.0874932531 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 10 |
Hb_012150_030 |
0.0878831079 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000340_150 |
0.0899860736 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 12 |
Hb_003053_110 |
0.0900900665 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005511_140 |
0.0907312821 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 14 |
Hb_006907_060 |
0.091558532 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002078_340 |
0.0945651916 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000941_100 |
0.0984032721 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 17 |
Hb_001014_070 |
0.0988895734 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 18 |
Hb_004109_260 |
0.0989931507 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 19 |
Hb_002631_240 |
0.0996266172 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 20 |
Hb_022378_010 |
0.1006592206 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |