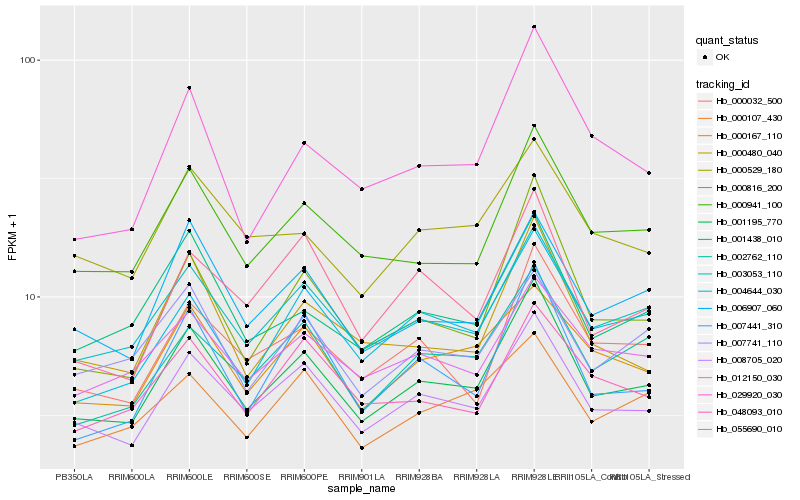
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_770 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 2 |
Hb_008705_020 |
0.0470348219 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_003053_110 |
0.0560570049 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000941_100 |
0.0638837933 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 5 |
Hb_000816_200 |
0.0688832009 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 6 |
Hb_006907_060 |
0.0727837818 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002762_110 |
0.0753184986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007441_310 |
0.076645506 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000480_040 |
0.0781303898 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 10 |
Hb_001438_010 |
0.078528016 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004644_030 |
0.079621748 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 12 |
Hb_000032_500 |
0.0803080466 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 13 |
Hb_000167_110 |
0.0812161188 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_012150_030 |
0.0848702076 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_029920_030 |
0.0859494936 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_048093_010 |
0.0868382292 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000529_180 |
0.087097647 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000107_430 |
0.0876050107 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 19 |
Hb_055690_010 |
0.0878122438 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007741_110 |
0.0880842274 |
- |
- |
- |