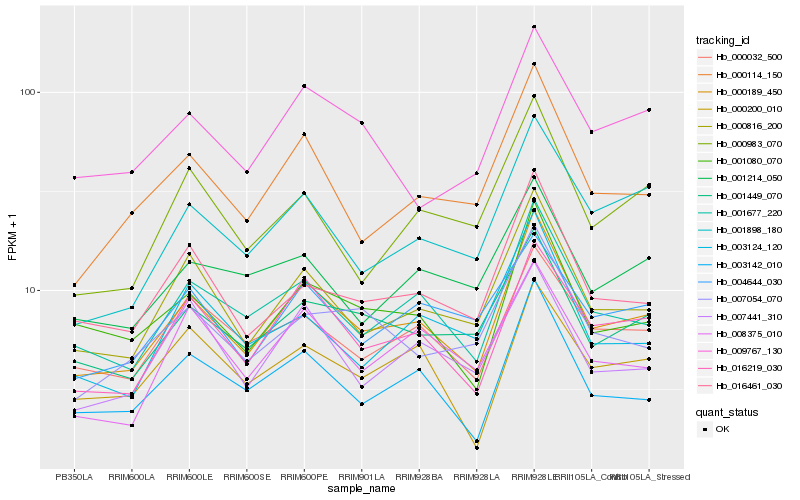
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_450 |
0.0 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001677_220 |
0.0727834423 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 3 |
Hb_000816_200 |
0.073528361 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 4 |
Hb_016219_030 |
0.0748377267 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_003142_010 |
0.0822954023 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 6 |
Hb_001080_070 |
0.085065257 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 7 |
Hb_000114_150 |
0.0896223815 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_001449_070 |
0.0918153769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_016461_030 |
0.0932768744 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_000032_500 |
0.0947355954 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 11 |
Hb_001898_180 |
0.0965473728 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000200_010 |
0.1059305389 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 13 |
Hb_009767_130 |
0.1076572938 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 14 |
Hb_008375_010 |
0.1077514729 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 15 |
Hb_007054_070 |
0.1082164851 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 16 |
Hb_000983_070 |
0.1082580344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007441_310 |
0.1086570264 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004644_030 |
0.109363568 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 19 |
Hb_003124_120 |
0.1101126827 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 20 |
Hb_001214_050 |
0.111368622 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |