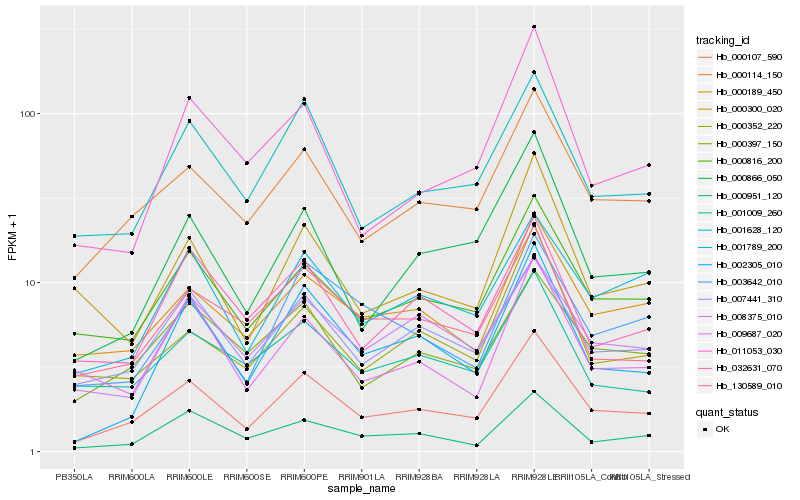
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_590 |
0.0 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 2 |
Hb_000114_150 |
0.0845703687 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_000816_200 |
0.1034760662 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 4 |
Hb_001628_120 |
0.1061931624 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 5 |
Hb_007441_310 |
0.1070313009 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002305_010 |
0.1085068594 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000866_050 |
0.1124932928 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_000189_450 |
0.1133168307 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000397_150 |
0.1190249184 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 10 |
Hb_000951_120 |
0.1193644915 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 11 |
Hb_001789_200 |
0.1205137596 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 12 |
Hb_130589_010 |
0.126019484 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_008375_010 |
0.1264092551 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 14 |
Hb_003642_010 |
0.1280127464 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_032631_070 |
0.1286125519 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000352_220 |
0.1287674675 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001009_260 |
0.1296312019 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000300_020 |
0.1304749683 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 19 |
Hb_009687_020 |
0.13072737 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 20 |
Hb_011053_030 |
0.1329165241 |
- |
- |
- |