| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
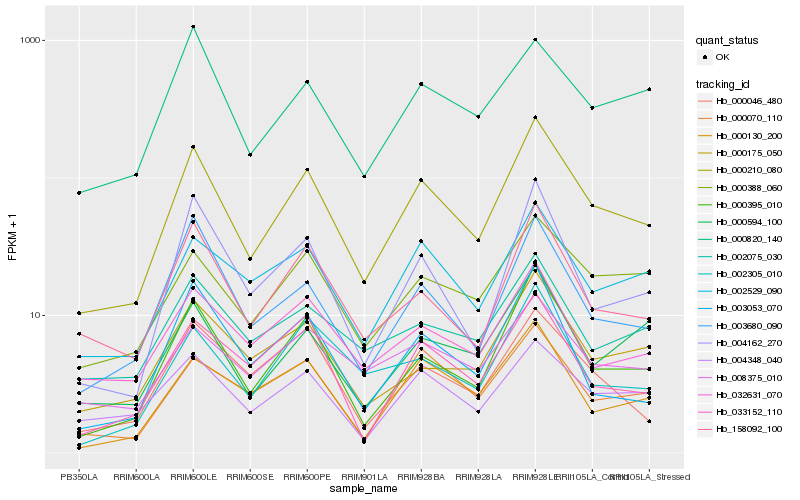
Hb_000210_080 |
0.0 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000395_010 |
0.0984967608 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 3 |
Hb_158092_100 |
0.117136262 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 4 |
Hb_002529_090 |
0.1219289974 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_000388_060 |
0.1234065777 |
- |
- |
fructokinase [Manihot esculenta] |
| 6 |
Hb_003680_090 |
0.1276027017 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004348_040 |
0.1276626349 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 8 |
Hb_004162_270 |
0.1287425946 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000175_050 |
0.1305416291 |
- |
- |
- |
| 10 |
Hb_000130_200 |
0.1318796368 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 11 |
Hb_000070_110 |
0.1337166865 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 12 |
Hb_032631_070 |
0.1337586361 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_000594_100 |
0.1353691421 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002305_010 |
0.1360472808 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_008375_010 |
0.1373716248 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 16 |
Hb_000046_480 |
0.1376652508 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 17 |
Hb_033152_110 |
0.1385277753 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 18 |
Hb_002075_030 |
0.1392441789 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003053_070 |
0.1402284564 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000820_140 |
0.1408927937 |
- |
- |
histone H4 [Zea mays] |