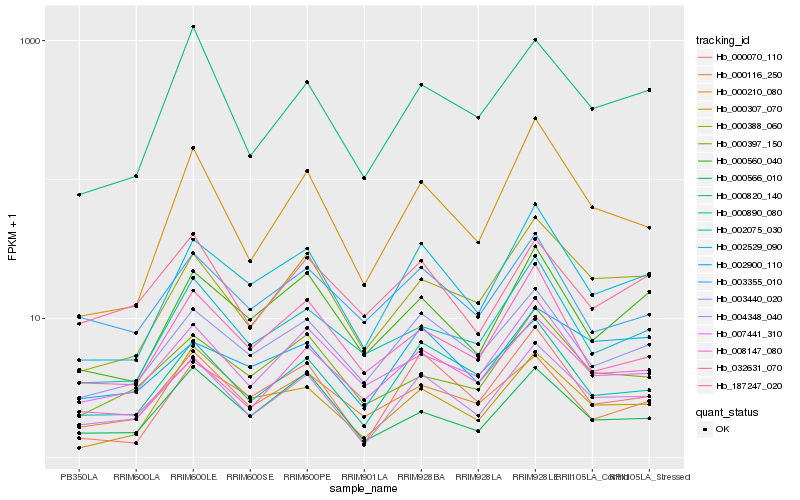
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004348_040 |
0.0 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 2 |
Hb_000890_080 |
0.1008415288 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 3 |
Hb_002529_090 |
0.1067511233 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_003440_020 |
0.1082726154 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000388_060 |
0.1153406899 |
- |
- |
fructokinase [Manihot esculenta] |
| 6 |
Hb_007441_310 |
0.1213226225 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000820_140 |
0.122774963 |
- |
- |
histone H4 [Zea mays] |
| 8 |
Hb_000210_080 |
0.1276626349 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_187247_020 |
0.1310181773 |
- |
- |
PREDICTED: uncharacterized protein LOC101220469 [Cucumis sativus] |
| 10 |
Hb_000397_150 |
0.1311964344 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 11 |
Hb_008147_080 |
0.1324306915 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000560_040 |
0.133710487 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_032631_070 |
0.1347359112 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_000116_250 |
0.134751846 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 15 |
Hb_000566_010 |
0.1349784762 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 16 |
Hb_000070_110 |
0.1352586007 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000307_070 |
0.1366674097 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_003355_010 |
0.1374878369 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 19 |
Hb_002900_110 |
0.1389823069 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_002075_030 |
0.1392906813 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |