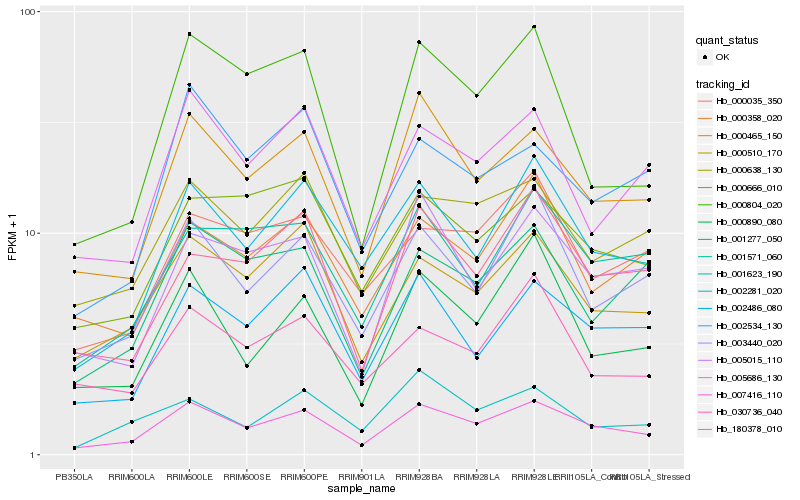
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_110 |
0.0 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000465_150 |
0.0791881039 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 3 |
Hb_002281_020 |
0.0949650044 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 4 |
Hb_000510_170 |
0.0958760026 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 5 |
Hb_003440_020 |
0.1016139601 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_002486_080 |
0.1026800432 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 7 |
Hb_000638_130 |
0.1089605747 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 8 |
Hb_005015_110 |
0.1095588803 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 9 |
Hb_000890_080 |
0.1128768118 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 10 |
Hb_005686_130 |
0.1129170711 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 11 |
Hb_001571_060 |
0.1144755244 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 12 |
Hb_180378_010 |
0.1155085374 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 13 |
Hb_000804_020 |
0.1161667672 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 14 |
Hb_000666_010 |
0.1165843528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001277_050 |
0.1182408818 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000358_020 |
0.1221928985 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 17 |
Hb_030736_040 |
0.1253924207 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002534_130 |
0.1260421922 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 19 |
Hb_001623_190 |
0.1269467426 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 20 |
Hb_000035_350 |
0.127006488 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |