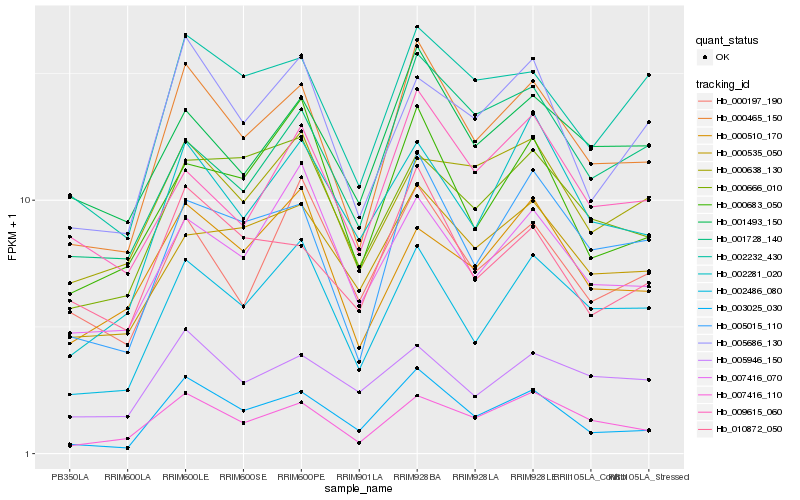
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_150 |
0.0 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 2 |
Hb_007416_110 |
0.0791881039 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005015_110 |
0.0799699184 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 4 |
Hb_002486_080 |
0.0918299879 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 5 |
Hb_003025_030 |
0.0976554062 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_002232_430 |
0.1001138516 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001493_150 |
0.1027972635 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 8 |
Hb_010872_050 |
0.1054107307 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 9 |
Hb_005686_130 |
0.1064716428 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 10 |
Hb_000535_050 |
0.1105524633 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 11 |
Hb_009615_060 |
0.1148618863 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 12 |
Hb_000666_010 |
0.1155940774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000638_130 |
0.1185727515 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 14 |
Hb_000510_170 |
0.1187822438 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 15 |
Hb_005946_150 |
0.1189086236 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 16 |
Hb_000197_190 |
0.1194064137 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_002281_020 |
0.1195205869 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 18 |
Hb_007416_070 |
0.1196211213 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 19 |
Hb_000683_050 |
0.1199497239 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 20 |
Hb_001728_140 |
0.120091898 |
- |
- |
- |